当前位置:
X-MOL 学术
›
Mol. Oral Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deletion of csn2 gene affects acid tolerance and exopolysaccharide synthesis in Streptococcus mutans.
Molecular Oral Microbiology ( IF 2.8 ) Pub Date : 2020-08-14 , DOI: 10.1111/omi.12308 Anqi Zhang 1 , Jiamin Chen 1 , Tao Gong 1 , Miao Lu 1 , Boyu Tang 1 , Xuedong Zhou 1 , Yuqing Li 1
Molecular Oral Microbiology ( IF 2.8 ) Pub Date : 2020-08-14 , DOI: 10.1111/omi.12308 Anqi Zhang 1 , Jiamin Chen 1 , Tao Gong 1 , Miao Lu 1 , Boyu Tang 1 , Xuedong Zhou 1 , Yuqing Li 1
Affiliation
|
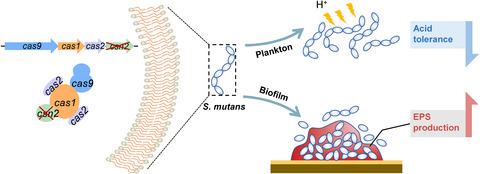
Csn2 is an important protein of the CRISPR‐Cas system. The physiological function of this protein and its regulatory role in Streptococcus mutans, as the primary causative agent of human dental caries, is still unclear. In this study, we investigated whether csn2 deletion would affect S. mutans physiology and virulence gene expression. We used microscopic imaging, acid killing assays, pH drop, biofilm formation, and exopolysaccharide (EPS) production tests to determine whether csn2 deletion influenced S. mutans colony morphology, acid tolerance/production, and glucan formation abilities. Comparisons were made between quantitative Real‐Time Polymerase Chain Reaction (qRT‐PCR) data from the UA159 and csn2 deletion strain to determine the impact of csn2 knockout on S. mutans gene expression. The results showed that deletion of S. mutans csn2 changed its colony morphotype and made it more sensitive to acid. The expression levels of aciduricity genes, including leuA, leuB, leuC, and leuD, were significantly down‐regulated. Acid adaptation restored the aciduricity of csn2 mutant and enhanced the ability to synthesize EPS. The expression levels of EPS synthesis‐related genes, including gtfC and gtfD, were significantly up‐regulated after acid adaptation. In summary, deletion of S. mutans csn2 exerted multiple effects on the virulence traits of this pathogen, including acid tolerance and EPS formation, and that these alterations could partially be attributed to changes in gene expression upon loss of csn2. Understanding the function of csn2 in S. mutans might lead to novel strategies to prevent or treat imbalances in oral microbiota that may favor diseases.
中文翻译:

csn2基因的缺失会影响变形链球菌的耐酸性和胞外多糖合成。
Csn2是CRISPR‐Cas系统的重要蛋白。尚不清楚该蛋白的生理功能及其在变形链球菌中的调节作用,而变形链球菌是人类龋齿的主要病因。在这项研究中,我们调查了csn2缺失是否会影响变形链球菌的生理和毒力基因表达。我们使用显微镜成像,杀酸测定,pH下降,生物膜形成和胞外多糖(EPS)生产测试来确定csn2缺失是否影响变形链球菌菌落形态,酸耐受性/生产和葡聚糖形成能力。对来自UA159和UA159的定量实时聚合酶链反应(qRT-PCR)数据进行了比较csn2缺失菌株,以确定csn2基因敲除对变形链球菌基因表达的影响。结果表明,变形链球菌 csn2的缺失改变了其菌落形态,并使其对酸更加敏感。包括leuA,leuB,leuC和leuD在内的酸性基因的表达水平明显下调。酸适应恢复了csn2突变体的酸性,并增强了合成EPS的能力。EPS合成相关基因(包括gtfC和gtfD)的表达水平酸适应后显着上调。总之,缺失变形链球菌 csn2对该病原体的毒力特性具有多种影响,包括耐酸性和EPS形成,这些改变可能部分归因于csn2缺失后基因表达的变化。了解csn2在变形链球菌中的功能可能会导致预防或治疗可能有利于疾病的口腔微生物群失衡的新策略。
更新日期:2020-09-26
中文翻译:

csn2基因的缺失会影响变形链球菌的耐酸性和胞外多糖合成。
Csn2是CRISPR‐Cas系统的重要蛋白。尚不清楚该蛋白的生理功能及其在变形链球菌中的调节作用,而变形链球菌是人类龋齿的主要病因。在这项研究中,我们调查了csn2缺失是否会影响变形链球菌的生理和毒力基因表达。我们使用显微镜成像,杀酸测定,pH下降,生物膜形成和胞外多糖(EPS)生产测试来确定csn2缺失是否影响变形链球菌菌落形态,酸耐受性/生产和葡聚糖形成能力。对来自UA159和UA159的定量实时聚合酶链反应(qRT-PCR)数据进行了比较csn2缺失菌株,以确定csn2基因敲除对变形链球菌基因表达的影响。结果表明,变形链球菌 csn2的缺失改变了其菌落形态,并使其对酸更加敏感。包括leuA,leuB,leuC和leuD在内的酸性基因的表达水平明显下调。酸适应恢复了csn2突变体的酸性,并增强了合成EPS的能力。EPS合成相关基因(包括gtfC和gtfD)的表达水平酸适应后显着上调。总之,缺失变形链球菌 csn2对该病原体的毒力特性具有多种影响,包括耐酸性和EPS形成,这些改变可能部分归因于csn2缺失后基因表达的变化。了解csn2在变形链球菌中的功能可能会导致预防或治疗可能有利于疾病的口腔微生物群失衡的新策略。







































 京公网安备 11010802027423号
京公网安备 11010802027423号