Journal of Proteomics ( IF 2.8 ) Pub Date : 2020-08-14 , DOI: 10.1016/j.jprot.2020.103940 Ming Chen 1 , Yi Li 1 , Houchen Lv 1 , Pengbin Yin 1 , Licheng Zhang 1 , Peifu Tang 1
|
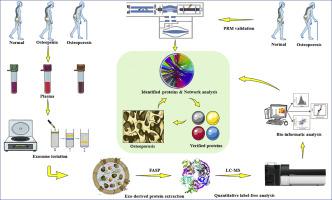
Alongside an aging population, osteoporosis has become increasingly common, representing a major public health problem. Human blood provides the predominant matrix for pathological targets underlining disease mechanisms. In the present study, the protein profiles of blood plasma exosomes from patients with osteoporosis, osteopenia, and those with normal bone mass were compared. The aim of the study was to search for potential novel diagnostic/therapeutic targets for further investigation in osteoporosis. A total of 60 participants were included from the PLAGH Hip Fracture Database. Quantitative proteomics was carried out to profile the plasma exosome derived proteins from patients diagnosed with osteoporosis, osteopenia, and normal bone mass, respectively. A Parallel reaction monitoring (PRM) analysis was further carried out to validate the identified proteins. Bio-informatics analyses including GO annotation and reverse engineering of gene regulatory networks analysis were applied in annotating the biological relevance of the identified proteins. Forty-five differentially expressed proteins were identified in the discovery dataset and four of them, PSMB9, AARS, PCBP2, and VSIR were further verified in a validation set. Based on the results, an exosomal-proteins index was constructed to classify individuals with osteoporosis from those without, an AUC of 0.805 (95% CI 0.620–0.926, p < 0.001) was achieved in classification performance assessment. Additionally, a reverse engineer of the regulatory network analysis identified and predicted the proteins which may interact with the four target proteins identified, providing references for further investigations into the pathological mechanisms of osteoporosis.
中文翻译:

定量蛋白质组学和逆向工程分析确定了与骨质疏松症相关的血浆外来体衍生蛋白标记。
随着人口老龄化,骨质疏松症变得越来越普遍,这是一个重大的公共卫生问题。人血为强调疾病机制的病理学靶标提供了主要基质。在本研究中,比较了患有骨质疏松症,骨质减少和正常骨量的患者血浆外泌体的蛋白质谱。该研究的目的是寻找潜在的新型诊断/治疗靶标,以进一步研究骨质疏松症。PLAGH髋部骨折数据库中共有60名参与者。进行了定量蛋白质组学分析,分别从诊断出患有骨质疏松症,骨质减少和正常骨量的患者的血浆外来体中提取出蛋白质。进一步进行了平行反应监测(PRM)分析以验证所鉴定的蛋白质。生物信息学分析包括GO注释和基因调控网络分析的逆向工程被用于注释已鉴定蛋白质的生物学相关性。在发现数据集中鉴定出了45种差异表达的蛋白质,其中的4种PSMB9,AARS,PCBP2和VSIR在验证集中进行了进一步验证。根据结果,构建了外泌体蛋白指数以将骨质疏松症患者与无骨质疏松症的患者进行分类,AUC为0.805(95%CI 0.620–0.926,在发现数据集中鉴定出了45种差异表达的蛋白质,其中的4种PSMB9,AARS,PCBP2和VSIR在验证集中进行了进一步验证。根据结果,构建了外泌体蛋白指数以将骨质疏松症患者与无骨质疏松症的患者进行分类,AUC为0.805(95%CI 0.620–0.926,在发现数据集中鉴定出45种差异表达的蛋白质,其中的4种PSMB9,AARS,PCBP2和VSIR在验证集中进一步验证。根据结果,构建了外泌体蛋白指数以将骨质疏松症患者与无骨质疏松症的患者进行分类,AUC为0.805(95%CI 0.620–0.926,p <0.001)在分类绩效评估中达到。此外,监管网络分析的逆向工程人员鉴定并预测了可能与鉴定出的四种靶蛋白相互作用的蛋白,为进一步研究骨质疏松症的病理机制提供了参考。











































 京公网安备 11010802027423号
京公网安备 11010802027423号