当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Latent Models of Molecular Dynamics Data: Automatic Order Parameter Generation for Peptide Fibrillization.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c05763 Nathaniel Charest 1 , Michael Tro 1 , Michael T Bowers 1 , Joan-Emma Shea 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c05763 Nathaniel Charest 1 , Michael Tro 1 , Michael T Bowers 1 , Joan-Emma Shea 1
Affiliation
|
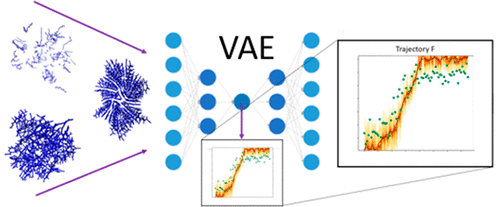
Variational autoencoders are artificial neural networks with the capability to reduce highly dimensional sets of data to smaller dimensional, latent representations. In this work, these models are applied to molecular dynamics simulations of the self-assembly of coarse-grained peptides to obtain a singled-valued order parameter for amyloid aggregation. This automatically learned order parameter is constructed by time-averaging the latent parametrizations of internal coordinate representations and compared to the nematic order parameter which is commonly used to study ordering of similar systems in literature. It is found that the latent space value provides more tailored insight into the aggregation mechanism’s details, correctly identifying fibril formation in instances where the nematic order parameter fails to do so. A means is provided by which the latent space value can be analyzed so that the major contributing internal coordinates are identified, allowing for a direct interpretation of the latent space order parameter in terms of the behavior of the system. The latent model is found to be an effective and convenient way of representing the data from the dynamic ensemble and provides a means of reducing the dimensionality of a system whose scale exceeds molecular systems so-far considered with similar tools. This bypasses a need for researcher speculation on what elements of a system best contribute to summarizing major transitions and suggests latent models are effective and insightful when applied to large systems with a diversity of complex behaviors.
中文翻译:

分子动力学数据的潜在模型:肽原纤化的自动顺序参数生成。
变体自动编码器是人工神经网络,具有将高维数据集减少为较小维,潜在表示的能力。在这项工作中,将这些模型应用于粗粒多肽自组装的分子动力学模拟,以获得淀粉样蛋白聚集的单值阶参数。该自动学习的顺序参数是通过对内部坐标表示形式的潜在参量进行时间平均来构造的,并与通常用于研究文献中类似系统的顺序的向列顺序参数进行比较。发现潜在空间值提供了对聚集机制细节的更量身定制的洞察力,在向列序参数未能做到的情况下正确地识别了原纤维形成。提供一种装置,通过该装置可以分析潜在空间值,从而识别出主要的内部坐标,从而可以根据系统的行为直接解释潜在空间顺序参数。发现潜在模型是表示动态集合中数据的一种有效且便捷的方式,并提供了一种减小系统规模的方法,该系统的规模超过了迄今为止使用类似工具考虑的分子系统。这就避免了研究人员对系统的哪些元素最有助于概括主要过渡的猜测,并建议当将潜在模型应用于具有多种复杂行为的大型系统时,它们是有效且有见地的。
更新日期:2020-09-18
中文翻译:

分子动力学数据的潜在模型:肽原纤化的自动顺序参数生成。
变体自动编码器是人工神经网络,具有将高维数据集减少为较小维,潜在表示的能力。在这项工作中,将这些模型应用于粗粒多肽自组装的分子动力学模拟,以获得淀粉样蛋白聚集的单值阶参数。该自动学习的顺序参数是通过对内部坐标表示形式的潜在参量进行时间平均来构造的,并与通常用于研究文献中类似系统的顺序的向列顺序参数进行比较。发现潜在空间值提供了对聚集机制细节的更量身定制的洞察力,在向列序参数未能做到的情况下正确地识别了原纤维形成。提供一种装置,通过该装置可以分析潜在空间值,从而识别出主要的内部坐标,从而可以根据系统的行为直接解释潜在空间顺序参数。发现潜在模型是表示动态集合中数据的一种有效且便捷的方式,并提供了一种减小系统规模的方法,该系统的规模超过了迄今为止使用类似工具考虑的分子系统。这就避免了研究人员对系统的哪些元素最有助于概括主要过渡的猜测,并建议当将潜在模型应用于具有多种复杂行为的大型系统时,它们是有效且有见地的。









































 京公网安备 11010802027423号
京公网安备 11010802027423号