当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Capturing Biologically Complex Tissue-Specific Membranes at Different Levels of Compositional Complexity.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c03368 Helgi I Ingólfsson 1 , Harsh Bhatia 2 , Talia Zeppelin 3 , W F Drew Bennett 1 , Kristy A Carpenter 4 , Pin-Chia Hsu 3 , Gautham Dharuman 1 , Peer-Timo Bremer 2, 5 , Birgit Schiøtt 3 , Felice C Lightstone 1 , Timothy S Carpenter 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c03368 Helgi I Ingólfsson 1 , Harsh Bhatia 2 , Talia Zeppelin 3 , W F Drew Bennett 1 , Kristy A Carpenter 4 , Pin-Chia Hsu 3 , Gautham Dharuman 1 , Peer-Timo Bremer 2, 5 , Birgit Schiøtt 3 , Felice C Lightstone 1 , Timothy S Carpenter 1
Affiliation
|
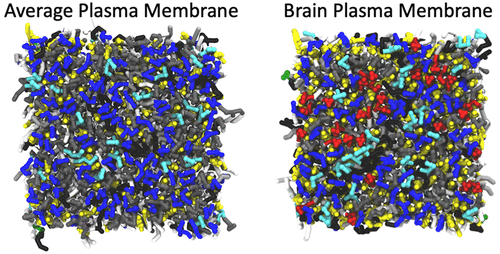
Plasma membranes (PMs) contain hundreds of different lipid species that contribute differently to overall bilayer properties. By modulation of these properties, membrane protein function can be affected. Furthermore, inhomogeneous lipid mixing and domains of lipid enrichment/depletion can sort proteins and provide optimal local environments. Recent coarse-grained (CG) Martini molecular dynamics efforts have provided glimpses into lipid organization of different PMs: an “Average” and a “Brain” PM. Their high complexity and large size require long simulations (∼80 μs) for proper sampling. Thus, these simulations are computationally taxing. This level of complexity is beyond the possibilities of all-atom simulations, raising the question—what complexity is needed for “realistic” bilayer properties? We constructed CG Martini PM models of varying complexity (63 down to 8 different lipids). Lipid tail saturations and headgroup combinations were kept as consistent as possible for the “tissues’” (Average/Brain) at three levels of compositional complexity. For each system, we analyzed membrane properties to evaluate which features can be retained at lower complexity and validate eight-component bilayers that can act as reliable mimetics for Average or Brain PMs. Systems of reduced complexity deliver a more robust and malleable tool for computational membrane studies and allow for equivalent all-atom simulations and experiments.
中文翻译:

在不同水平的组成复杂性上捕获生物复杂的组织特定膜。
质膜(PM)包含数百种不同的脂质种类,它们对整体双层特性的贡献不同。通过调节这些特性,可以影响膜蛋白的功能。此外,不均匀的脂质混合和脂质富集/耗尽的区域可以分选蛋白质并提供最佳的局部环境。最近的粗颗粒(CG)马提尼分子动力学研究成果使人们可以窥见不同PM的脂质组织:“平均”和“脑” PM。它们的高复杂度和大尺寸要求长时间的仿真(〜80μs)以进行正确的采样。因此,这些模拟在计算上很费力。这种复杂程度超出了所有原子模拟的可能性,提出了一个问题:“现实的”双层属性需要什么复杂性?我们构建了复杂程度各异的CG Martini PM模型(63到8种不同的脂质)。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。
更新日期:2020-09-10
中文翻译:

在不同水平的组成复杂性上捕获生物复杂的组织特定膜。
质膜(PM)包含数百种不同的脂质种类,它们对整体双层特性的贡献不同。通过调节这些特性,可以影响膜蛋白的功能。此外,不均匀的脂质混合和脂质富集/耗尽的区域可以分选蛋白质并提供最佳的局部环境。最近的粗颗粒(CG)马提尼分子动力学研究成果使人们可以窥见不同PM的脂质组织:“平均”和“脑” PM。它们的高复杂度和大尺寸要求长时间的仿真(〜80μs)以进行正确的采样。因此,这些模拟在计算上很费力。这种复杂程度超出了所有原子模拟的可能性,提出了一个问题:“现实的”双层属性需要什么复杂性?我们构建了复杂程度各异的CG Martini PM模型(63到8种不同的脂质)。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。脂质尾部饱和度和头基组合对于“组织”(平均/大脑)在三个组成复杂度上保持尽可能一致。对于每个系统,我们分析了膜的性能,以评估哪些功能可以较低的复杂度保留下来,并验证了八组分双层膜,它们可以用作平均PM或脑PM的可靠模拟物。复杂性降低的系统为计算膜研究提供了更强大且更具延展性的工具,并允许进行等效的全原子模拟和实验。











































 京公网安备 11010802027423号
京公网安备 11010802027423号