当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
ADD Force Field for Sugars and Polyols: Predicting the Additivity of Protein-Osmolyte Interaction.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c05345 Andrea Arsiccio 1 , Pritam Ganguly 1 , Lorenzo La Cortiglia 2 , Joan-Emma Shea 1, 3 , Roberto Pisano 2
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-08-11 , DOI: 10.1021/acs.jpcb.0c05345 Andrea Arsiccio 1 , Pritam Ganguly 1 , Lorenzo La Cortiglia 2 , Joan-Emma Shea 1, 3 , Roberto Pisano 2
Affiliation
|
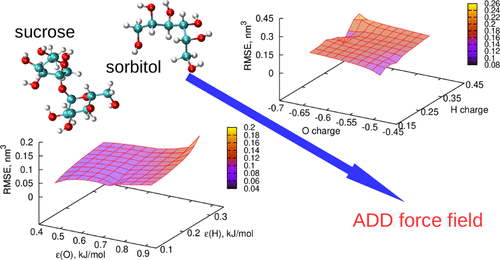
The protein–osmolyte interaction has been shown experimentally to follow an additive construct, where the individual osmolyte–backbone and osmolyte–side-chain interactions contribute to the overall conformational stability of proteins. Here, we computationally reconstruct this additive relation using molecular dynamics simulations, focusing on sugars and polyols, including sucrose and sorbitol, as model osmolytes. A new set of parameters (ADD) is developed for this purpose, using the individual Kirkwood–Buff integrals for sugar–backbone and sugar–side-chain interactions as target experimental data. We show that the ADD parameters can reproduce the additivity of protein–sugar interactions and correctly predict sucrose and sorbitol self-association and their interaction with water. The accurate description of the separate osmolyte–backbone and osmolyte–side-chain contributions also automatically translates into a good prediction of preferential exclusion from the surface of ribonuclease A and α-chymotrypsinogen A. The description of sugar polarity is improved compared to previous force fields, resulting in closer agreement with the experimental data and better compatibility with charged groups, such as the guanidinium moiety. The ADD parameters are developed in combination with the CHARMM36m force field for proteins, but good compatibility is also observed with the AMBER 99SB-ILDN and the OPLS-AA force fields. Overall, exploiting the additivity of protein–osmolyte interactions is a promising approach for the development of new force fields.
中文翻译:

糖和多元醇的添加力场:预测蛋白质-渗透物相互作用的加性。
实验证明,蛋白质与渗透物的相互作用遵循可加性结构,其中渗透物与骨干之间的相互作用以及渗透物与侧链之间的相互作用对蛋白质的整体构象稳定性有贡献。在这里,我们使用分子动力学模拟,以糖和多元醇(包括蔗糖和山梨糖醇)为模型渗透物,以计算方式重建了这种加性关系。为此,开发了一组新的参数(ADD),使用了糖-主链和糖-侧链相互作用的单个Kirkwood-Buff积分作为目标实验数据。我们表明,ADD参数可以再现蛋白质与糖相互作用的加性,并正确预测蔗糖和山梨糖醇的自缔合以及它们与水的相互作用。对渗透压-主链和渗透压-侧链贡献的准确描述也自动转化为对核糖核酸酶A和α-胰凝乳蛋白酶原A表面优先排除的良好预测。与以前的力场相比,糖极性的描述得到了改进,从而与实验数据更加吻合,并与带电基团(例如胍基部分)具有更好的相容性。与蛋白质的CHARMM36m力场结合开发了ADD参数,但是与AMBER 99SB-ILDN和OPLS-AA力场也观察到了良好的兼容性。总的来说,利用蛋白质-渗透物相互作用的可加性是发展新的力场的有前途的方法。
更新日期:2020-09-10
中文翻译:

糖和多元醇的添加力场:预测蛋白质-渗透物相互作用的加性。
实验证明,蛋白质与渗透物的相互作用遵循可加性结构,其中渗透物与骨干之间的相互作用以及渗透物与侧链之间的相互作用对蛋白质的整体构象稳定性有贡献。在这里,我们使用分子动力学模拟,以糖和多元醇(包括蔗糖和山梨糖醇)为模型渗透物,以计算方式重建了这种加性关系。为此,开发了一组新的参数(ADD),使用了糖-主链和糖-侧链相互作用的单个Kirkwood-Buff积分作为目标实验数据。我们表明,ADD参数可以再现蛋白质与糖相互作用的加性,并正确预测蔗糖和山梨糖醇的自缔合以及它们与水的相互作用。对渗透压-主链和渗透压-侧链贡献的准确描述也自动转化为对核糖核酸酶A和α-胰凝乳蛋白酶原A表面优先排除的良好预测。与以前的力场相比,糖极性的描述得到了改进,从而与实验数据更加吻合,并与带电基团(例如胍基部分)具有更好的相容性。与蛋白质的CHARMM36m力场结合开发了ADD参数,但是与AMBER 99SB-ILDN和OPLS-AA力场也观察到了良好的兼容性。总的来说,利用蛋白质-渗透物相互作用的可加性是发展新的力场的有前途的方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号