Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Predicting the Real‐Valued Inter‐Residue Distances for Proteins
Advanced Science ( IF 14.3 ) Pub Date : 2020-08-10 , DOI: 10.1002/advs.202001314 Wenze Ding 1, 2 , Haipeng Gong 1, 2
Advanced Science ( IF 14.3 ) Pub Date : 2020-08-10 , DOI: 10.1002/advs.202001314 Wenze Ding 1, 2 , Haipeng Gong 1, 2
Affiliation
|
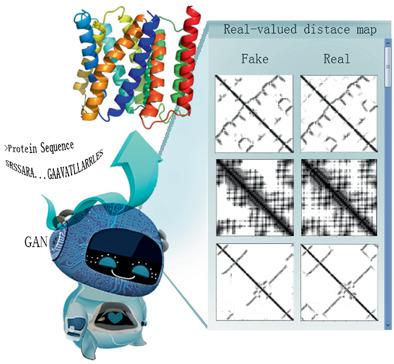
Predicting protein structure from the amino acid sequence has been a challenge with theoretical and practical significance in biophysics. Despite the recent progresses elicited by improved inter‐residue contact prediction, contact‐based structure prediction has gradually reached the performance ceiling. New methods have been proposed to predict the inter‐residue distance, but unanimously by simplifying the real‐valued distance prediction into a multiclass classification problem. Here, a lightweight regression‐based distance prediction method is shown, which adopts the generative adversarial network to capture the delicate geometric relationship between residue pairs and thus could predict the continuous, real‐valued inter‐residue distance rapidly and satisfactorily. The predicted residue distance map allows quick structure modeling by the CNS suite, and the constructed models approach the same level of quality as the other state‐of‐the‐art protein structure prediction methods when tested on CASP13 targets. Moreover, this method can be used directly for the structure prediction of membrane proteins without transfer learning.
中文翻译:

预测蛋白质的实值残基间距离
从氨基酸序列预测蛋白质结构一直是生物物理学中具有理论和实践意义的挑战。尽管最近通过改进残基间接触预测取得了进展,但基于接触的结构预测已逐渐达到性能上限。人们提出了新的方法来预测残基间距离,但一致将实值距离预测简化为多类分类问题。这里,提出了一种基于轻量级回归的距离预测方法,该方法采用生成对抗网络来捕获残基对之间微妙的几何关系,从而可以快速且令人满意地预测连续的、实值的残基间距离。预测的残基距离图允许 CNS 套件进行快速结构建模,并且在 CASP13 靶标上进行测试时,构建的模型与其他最先进的蛋白质结构预测方法具有相同的质量水平。而且,该方法可以直接用于膜蛋白的结构预测,无需迁移学习。
更新日期:2020-10-07
中文翻译:

预测蛋白质的实值残基间距离
从氨基酸序列预测蛋白质结构一直是生物物理学中具有理论和实践意义的挑战。尽管最近通过改进残基间接触预测取得了进展,但基于接触的结构预测已逐渐达到性能上限。人们提出了新的方法来预测残基间距离,但一致将实值距离预测简化为多类分类问题。这里,提出了一种基于轻量级回归的距离预测方法,该方法采用生成对抗网络来捕获残基对之间微妙的几何关系,从而可以快速且令人满意地预测连续的、实值的残基间距离。预测的残基距离图允许 CNS 套件进行快速结构建模,并且在 CASP13 靶标上进行测试时,构建的模型与其他最先进的蛋白质结构预测方法具有相同的质量水平。而且,该方法可以直接用于膜蛋白的结构预测,无需迁移学习。











































 京公网安备 11010802027423号
京公网安备 11010802027423号