Cell Chemical Biology ( IF 6.6 ) Pub Date : 2020-08-11 , DOI: 10.1016/j.chembiol.2020.07.019 Julie M Garlick 1 , Anna K Mapp 2
|
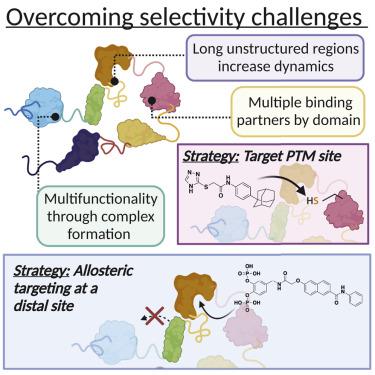
Dynamic proteins perform critical roles in cellular machines, including those that control proteostasis, transcription, translation, and signaling. Thus, dynamic proteins are prime candidates for chemical probe and drug discovery but difficult targets because they do not conform to classical rules of design and screening. Selectivity is pivotal for candidate probe molecules due to the extensive interaction network of these dynamic hubs. Recognition that the traditional rules of probe discovery are not necessarily applicable to dynamic proteins and their complexes, as well as technological advances in screening, have produced remarkable results in the last 2–4 years. Particularly notable are the improvements in target selectivity for small-molecule modulators of dynamic proteins, especially with techniques that increase the discovery likelihood of allosteric regulatory mechanisms. We focus on approaches to small-molecule screening that appear to be more suitable for highly dynamic targets and have the potential to streamline identification of selective modulators.
中文翻译:

动态蛋白质复合物的选择性调节。
动态蛋白质在细胞机器中发挥着关键作用,包括控制蛋白质稳态、转录、翻译和信号传导的蛋白质。因此,动态蛋白质是化学探针和药物发现的主要候选者,但却是困难的目标,因为它们不符合设计和筛选的经典规则。由于这些动态中心的广泛相互作用网络,选择性对于候选探针分子至关重要。人们认识到探针发现的传统规则不一定适用于动态蛋白质及其复合物,以及筛选技术的进步,在过去 2-4 年中取得了显着的成果。特别值得注意的是动态蛋白小分子调节剂的靶点选择性的提高,特别是通过增加变构调节机制发现可能性的技术。我们专注于小分子筛选方法,这些方法似乎更适合高动态靶标,并且有可能简化选择性调节剂的识别。









































 京公网安备 11010802027423号
京公网安备 11010802027423号