Medicinal Chemistry ( IF 1.9 ) Pub Date : 2020-07-31 , DOI: 10.2174/1573406415666191004125551 Zi-Mei Zhang 1 , Zheng-Xing Guan 1 , Fang Wang 1 , Dan Zhang 1 , Hui Ding 1
|
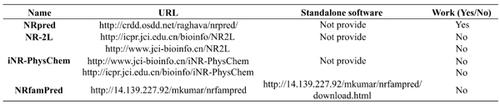
Nuclear receptors (NRs) are a superfamily of ligand-dependent transcription factors that are closely related to cell development, differentiation, reproduction, homeostasis, and metabolism. According to the alignments of the conserved domains, NRs are classified and assigned the following seven subfamilies or eight subfamilies: (1) NR1: thyroid hormone like (thyroid hormone, retinoic acid, RAR-related orphan receptor, peroxisome proliferator activated, vitamin D3- like), (2) NR2: HNF4-like (hepatocyte nuclear factor 4, retinoic acid X, tailless-like, COUP-TFlike, USP), (3) NR3: estrogen-like (estrogen, estrogen-related, glucocorticoid-like), (4) NR4: nerve growth factor IB-like (NGFI-B-like), (5) NR5: fushi tarazu-F1 like (fushi tarazu-F1 like), (6) NR6: germ cell nuclear factor like (germ cell nuclear factor), and (7) NR0: knirps like (knirps, knirpsrelated, embryonic gonad protein, ODR7, trithorax) and DAX like (DAX, SHP), or dividing NR0 into (7) NR7: knirps like and (8) NR8: DAX like. Different NRs families have different structural features and functions. Since the function of a NR is closely correlated with which subfamily it belongs to, it is highly desirable to identify NRs and their subfamilies rapidly and effectively. The knowledge acquired is essential for a proper understanding of normal and abnormal cellular mechanisms. With the advent of the post-genomics era, huge amounts of sequence-known proteins have increased explosively. Conventional methods for accurately classifying the family of NRs are experimental means with high cost and low efficiency. Therefore, it has created a greater need for bioinformatics tools to effectively recognize NRs and their subfamilies for the purpose of understanding their biological function. In this review, we summarized the application of machine learning methods in the prediction of NRs from different aspects. We hope that this review will provide a reference for further research on the classification of NRs and their families.
中文翻译:

机器学习方法在预测核受体及其家族中的应用
核受体(NRs)是配体依赖性转录因子的超家族,与细胞发育,分化,繁殖,体内稳态和代谢密切相关。根据保守域的排列方式,对NRs进行分类并分配了以下七个亚家族或八个亚家族:(1)NR1:甲状腺激素样(甲状腺激素,视黄酸,RAR相关的孤儿受体,过氧化物酶体增殖物激活,维生素D3- ),(2)NR2:HNF4样(肝细胞核因子4,视黄酸X,无尾状,COUP-TF样,USP),(3)NR3:雌激素样(雌激素,雌激素相关,糖皮质激素样) ),(4)NR4:神经生长因子IB样(NGFI-B样),(5)NR5:富士tarazu-F1样(fushi tarazu-F1样),(6)NR6:生殖细胞核因子如( (7)NR0:类似(knirps,与knirps相关的胚胎性腺蛋白,ODR7,trithorax)和DAX(DAX,SHP),或将NR0分为(7)NR7:knirps样和(8)NR8:DAX样。不同的NR系列具有不同的结构特征和功能。由于NR的功能与其所属的亚家族密切相关,因此迫切需要快速,有效地鉴定NR及其亚家族。获得的知识对于正确理解正常和异常细胞机制至关重要。随着后基因组学时代的到来,大量的序列已知蛋白呈爆炸性增长。准确分类NRs家族的常规方法是高成本和低效率的实验手段。因此,为了更好地了解NR及其亚家族,人们越来越需要生物信息学工具来有效识别NR及其亚家族。在这篇综述中,我们从不同方面总结了机器学习方法在NR预测中的应用。我们希望这次审查能为NRs及其家族分类的进一步研究提供参考。











































 京公网安备 11010802027423号
京公网安备 11010802027423号