Medicinal Chemistry ( IF 1.9 ) Pub Date : 2020-07-31 , DOI: 10.2174/1573406415666191004135450 Yuchi Zhang 1 , Xinyu Wu 2 , Cong Zhao 3 , Kai Li 4 , Yi Zheng 5 , Jing Zhao 1 , Pengling Ge 1
|
Background: Molecular characterization of insulin resistance, a growing health issue worldwide, will help to develop novel strategies and accurate biomarkers for disease diagnosis and treatment.
Objective: Integrative analysis of gene expression profiling and gene regulatory network was exploited to identify potential biomarkers early in the development of insulin resistance.
Methods: RNA was isolated from livers of animals at three weeks of age, and whole-genome expression profiling was performed and analyzed with Agilent mouse 4×44K microarrays. Differentially expressed genes were subsequently validated by qRT-PCR. Functional characterizations of genes and their interactions were performed by Gene Ontology (GO) analysis and gene regulatory network (GRN) analysis.
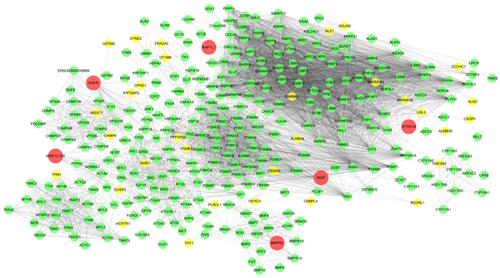
Results: A total of 197 genes were found to be differentially expressed by fold change ≥2 and P < 0.05 in BKS-db +/+ mice relative to sex and age-matched controls. Functional analysis suggested that these differentially expressed genes were enriched in the regulation of phosphorylation and generation of precursor metabolites which are closely associated with insulin resistance. Then a gene regulatory network associated with insulin resistance (IRGRN) was constructed by integration of these differentially expressed genes and known human protein-protein interaction network. The principal component analysis demonstrated that 67 genes in IRGRN could clearly distinguish insulin resistance from the non-disease state. Some of these candidate genes were further experimentally validated by qRT-PCR, highlighting the predictive role as biomarkers in insulin resistance.
Conclusion: Our study provides new insight into the pathogenesis and treatment of insulin resistance and also reveals potential novel molecular targets and diagnostic biomarkers for insulin resistance.
中文翻译:

全基因组表达谱和调控网络的综合分析确定了瘦素受体缺陷型小鼠胰岛素抵抗的新型生物标记。
背景:胰岛素抵抗的分子表征是全球范围内日益严重的健康问题,将有助于开发新颖的策略和准确的生物标志物,用于疾病的诊断和治疗。
目的:利用基因表达谱分析和基因调控网络的综合分析,在胰岛素抵抗发展早期发现潜在的生物标志物。
方法:从三周大的动物肝脏中分离RNA,进行全基因组表达谱分析,并使用Agilent小鼠4×44K芯片进行分析。随后通过qRT-PCR验证差异表达的基因。基因的功能表征及其相互作用通过基因本体论(GO)分析和基因调控网络(GRN)分析进行。
结果:与性别和年龄相匹配的对照组相比,在BKS-db + / +小鼠中,共有197个基因的差异倍数≥2和P <0.05差异表达。功能分析表明,这些差异表达的基因在磷酸化的调节和与胰岛素抵抗密切相关的前体代谢产物的产生中富集。然后,通过整合这些差异表达的基因和已知的人蛋白质-蛋白质相互作用网络,构建与胰岛素抵抗相关的基因调控网络(IRGRN)。主成分分析表明,IRGRN中的67个基因可以清楚地区分胰岛素抵抗与非疾病状态。其中一些候选基因通过qRT-PCR进一步进行了实验验证,
结论:我们的研究为胰岛素抵抗的发病机理和治疗提供了新的见解,并揭示了潜在的新型分子靶标和胰岛素抵抗的诊断生物标志物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号