当前位置:
X-MOL 学术
›
Curr. Genomics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Metagenomic exploration of plastic degrading microbes for biotechnological application
Current Genomics ( IF 1.8 ) Pub Date : 2020-08-08 , DOI: 10.2174/1389202921999200525155711 Jyotika Purohit 1 , Anirudha Chattopadhyay 1 , Basavaraj Teli 2
Current Genomics ( IF 1.8 ) Pub Date : 2020-08-08 , DOI: 10.2174/1389202921999200525155711 Jyotika Purohit 1 , Anirudha Chattopadhyay 1 , Basavaraj Teli 2
Affiliation
|
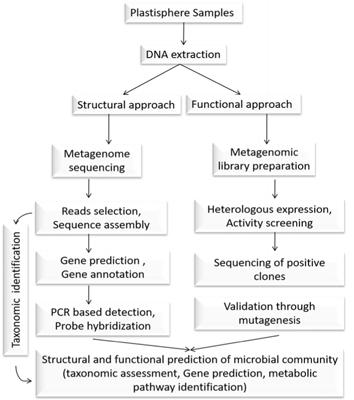
Since the last few decades, the promiscuous and uncontrolled use of plastics led to the accumulation of millions of tons of plastic waste in the terrestrial and marine environment. It elevated the risk of environmental pollution and climate change. The concern arises more due to the reckless and unscientific disposal of plastics containing high molecular weight polymers, viz., polystyrene, polyamide, polyvinylchloride, polypropylene, polyurethane, and polyethylene, etc. which are very difficult to degrade. Thus, the focus is now paid to search for efficient, eco-friendly, low-cost waste management technology. Of them, degradation of non-degradable synthetic polymer using diverse microbial agents, viz., bacteria, fungi, and other extremophiles become an emerging option. So far, very few microbial agents and their secreted enzymes have been identified and characterized for plastic degradation, but with low efficiency. It might be due to the predominance of uncultured microbial species, which consequently remain unexplored from the respective plastic degrading milieu. To overcome this problem, metagenomic analysis of microbial population engaged in the plastic biodegradation is advisable to decipher the microbial community structure and to predict their biodegradation potential in situ. Advancements in sequencing technologies and bioinformatics analysis allow the rapid metagenome screening that helps in the identification of total microbial community and also opens up the scope for mining genes or enzymes (hydrolases, laccase, etc.) engaged in polymer degradation. Further, the extraction of the core microbial population and their adaptation, fitness, and survivability can also be deciphered through comparative metagenomic study. It will help to engineer the microbial community and their metabolic activity to speed up the degradation process.
中文翻译:

用于生物技术应用的塑料降解微生物的宏基因组探索
自过去几十年以来,塑料的混杂和不受控制的使用导致了数百万吨塑料废物在陆地和海洋环境中的积累。它增加了环境污染和气候变化的风险。更令人担忧的是,对含有高分子量聚合物的塑料(即聚苯乙烯、聚酰胺、聚氯乙烯、聚丙烯、聚氨酯和聚乙烯等)的处理不计后果和不科学,这些聚合物很难降解。因此,现在的重点是寻找高效、环保、低成本的废物管理技术。其中,使用多种微生物制剂(即细菌、真菌和其他极端微生物)降解不可降解的合成聚合物成为一种新兴选择。迄今为止,很少有微生物及其分泌的酶被鉴定和表征用于塑料降解,但效率低下。这可能是由于未培养的微生物物种占主导地位,因此仍未从各自的塑料降解环境中进行探索。为了克服这个问题,建议对参与塑料生物降解的微生物种群进行宏基因组分析,以破译微生物群落结构并预测其原位生物降解潜力。测序技术和生物信息学分析的进步允许快速宏基因组筛选,有助于识别总微生物群落,并为挖掘参与聚合物降解的基因或酶(水解酶、漆酶等)开辟了空间。更多,核心微生物种群的提取及其适应性、适应性和生存能力也可以通过比较宏基因组研究来破译。它将有助于设计微生物群落及其代谢活动,以加快降解过程。
更新日期:2020-08-08
中文翻译:

用于生物技术应用的塑料降解微生物的宏基因组探索
自过去几十年以来,塑料的混杂和不受控制的使用导致了数百万吨塑料废物在陆地和海洋环境中的积累。它增加了环境污染和气候变化的风险。更令人担忧的是,对含有高分子量聚合物的塑料(即聚苯乙烯、聚酰胺、聚氯乙烯、聚丙烯、聚氨酯和聚乙烯等)的处理不计后果和不科学,这些聚合物很难降解。因此,现在的重点是寻找高效、环保、低成本的废物管理技术。其中,使用多种微生物制剂(即细菌、真菌和其他极端微生物)降解不可降解的合成聚合物成为一种新兴选择。迄今为止,很少有微生物及其分泌的酶被鉴定和表征用于塑料降解,但效率低下。这可能是由于未培养的微生物物种占主导地位,因此仍未从各自的塑料降解环境中进行探索。为了克服这个问题,建议对参与塑料生物降解的微生物种群进行宏基因组分析,以破译微生物群落结构并预测其原位生物降解潜力。测序技术和生物信息学分析的进步允许快速宏基因组筛选,有助于识别总微生物群落,并为挖掘参与聚合物降解的基因或酶(水解酶、漆酶等)开辟了空间。更多,核心微生物种群的提取及其适应性、适应性和生存能力也可以通过比较宏基因组研究来破译。它将有助于设计微生物群落及其代谢活动,以加快降解过程。











































 京公网安备 11010802027423号
京公网安备 11010802027423号