当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An Improved Intracellular Synthetic Lipidation-Induced Plasma Membrane Anchoring System for SNAP-Tag Fusion Proteins.
Biochemistry ( IF 2.9 ) Pub Date : 2020-08-10 , DOI: 10.1021/acs.biochem.0c00410 Tatsuyuki Yoshii 1, 2 , Kai Tahara 1 , Sachio Suzuki 3 , Yuka Hatano 1 , Keiko Kuwata 4 , Shinya Tsukiji 1, 3
Biochemistry ( IF 2.9 ) Pub Date : 2020-08-10 , DOI: 10.1021/acs.biochem.0c00410 Tatsuyuki Yoshii 1, 2 , Kai Tahara 1 , Sachio Suzuki 3 , Yuka Hatano 1 , Keiko Kuwata 4 , Shinya Tsukiji 1, 3
Affiliation
|
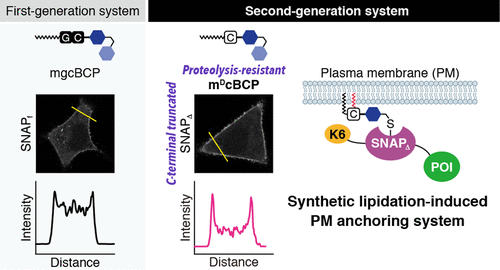
The ability to chemically introduce lipid modifications to specific intracellular protein targets would enable the conditional control of protein localization and activity in living cells. We recently developed a chemical–genetic approach in which an engineered SNAP-tag fusion protein can be rapidly relocated and anchored from the cytoplasm to the plasma membrane (PM) upon post-translational covalent lipopeptide conjugation in cells. However, the first-generation system achieved only low to moderate protein anchoring (recruiting) efficiencies and lacked wide applicability. Herein, we describe the rational design of an improved system for intracellular synthetic lipidation-induced PM anchoring of SNAP-tag fusion proteins. In the new system, the SNAPf protein engineered to contain an N-terminal hexalysine (K6) sequence and a C-terminal 10-amino acid deletion, termed K6-SNAPΔ, is fused to a protein of interest. In addition, a SNAP-tag substrate containing a metabolic-resistant myristoyl-DCys lipopeptidomimetic, called mDcBCP, is used as a cell-permeable chemical probe for intracellular SNAP-tag lipidation. The use of this combination allows significantly improved conditional PM anchoring of SNAP-tag fusion proteins. This second-generation system was applied to activate various signaling proteins, including Tiam1, cRaf, PI3K, and Sos, upon synthetic lipidation-induced PM anchoring/recruitment, offering a new and useful research tool in chemical biology and synthetic biology.
中文翻译:

SNAP标签融合蛋白的改进的细胞内合成脂质诱导的质膜锚定系统。
化学地将脂质修饰引入特定细胞内蛋白质靶标的能力将能够有条件地控制活细胞中蛋白质的定位和活性。我们最近开发了一种化学遗传方法,其中工程化的SNAP-tag融合蛋白可在细胞中翻译后共价脂肽缀合后迅速重定位并从细胞质锚定到质膜(PM)。但是,第一代系统仅实现了低到中等的蛋白质锚定(招募)效率,并且缺乏广泛的适用性。在这里,我们描述了细胞内合成脂质化诱导的SNAP标签融合蛋白的PM锚定的改良系统的合理设计。在新系统中,SNAP f蛋白工程改造以含有N-末端hexalysine(K6)序列和C端10个氨基酸的缺失,称作K6-SNAP Δ,融合到感兴趣的蛋白质。另外,包含抗代谢肉豆蔻酰基-D Cys脂肽模拟物的SNAP标签底物,称为m D cBCP,用作细胞内SNAP标签脂质化的细胞可渗透化学探针。这种组合的使用可以显着改善SNAP标签融合蛋白的条件PM锚定。该第二代系统在合成脂化诱导的PM锚定/招聘后,被用于激活各种信号蛋白,包括Tiam1,cRaf,PI3K和Sos,从而为化学生物学和合成生物学提供了一种新的有用的研究工具。
更新日期:2020-08-25
中文翻译:

SNAP标签融合蛋白的改进的细胞内合成脂质诱导的质膜锚定系统。
化学地将脂质修饰引入特定细胞内蛋白质靶标的能力将能够有条件地控制活细胞中蛋白质的定位和活性。我们最近开发了一种化学遗传方法,其中工程化的SNAP-tag融合蛋白可在细胞中翻译后共价脂肽缀合后迅速重定位并从细胞质锚定到质膜(PM)。但是,第一代系统仅实现了低到中等的蛋白质锚定(招募)效率,并且缺乏广泛的适用性。在这里,我们描述了细胞内合成脂质化诱导的SNAP标签融合蛋白的PM锚定的改良系统的合理设计。在新系统中,SNAP f蛋白工程改造以含有N-末端hexalysine(K6)序列和C端10个氨基酸的缺失,称作K6-SNAP Δ,融合到感兴趣的蛋白质。另外,包含抗代谢肉豆蔻酰基-D Cys脂肽模拟物的SNAP标签底物,称为m D cBCP,用作细胞内SNAP标签脂质化的细胞可渗透化学探针。这种组合的使用可以显着改善SNAP标签融合蛋白的条件PM锚定。该第二代系统在合成脂化诱导的PM锚定/招聘后,被用于激活各种信号蛋白,包括Tiam1,cRaf,PI3K和Sos,从而为化学生物学和合成生物学提供了一种新的有用的研究工具。



























 京公网安备 11010802027423号
京公网安备 11010802027423号