Plant Physiology ( IF 6.5 ) Pub Date : 2020-10-01 , DOI: 10.1104/pp.20.00865 Robert G Orr 1 , Stephen J Foley 2 , Catherine Sherman 1, 3 , Isidro Abreu 4 , Giulia Galotto 1 , Boyuan Liu 1 , Manuel González-Guerrero 4 , Luis Vidali 2, 5
|
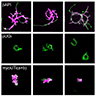
RNA interference (RNAi) enables flexible and dynamic interrogation of entire gene families or essential genes without the need for exogenous proteins, unlike CRISPR-Cas technology. Unfortunately, isolation of plants undergoing potent gene silencing requires laborious design, visual screening, and physical separation for downstream characterization. Here, we developed an adenine phosphoribosyltransferase (APT)-based RNAi technology (APTi) in Physcomitrella patens that improves upon the multiple limitations of current RNAi techniques. APTi exploits the prosurvival output of transiently silencing APT in the presence of 2-fluoroadenine, thereby establishing survival itself as a reporter of RNAi. To maximize the silencing efficacy of gene targets, we created vectors that facilitate insertion of any gene target sequence in tandem with the APT silencing motif. We tested the efficacy of APTi with two gene families, the actin-dependent motor, myosin XI (a,b), and the putative chitin receptor Lyk5 (a,b,c). The APTi approach resulted in a homogenous population of transient P. patens mutants specific for our gene targets with zero surviving background plants within 8 d. The observed mutants directly corresponded to a maximal 93% reduction of myosin XI protein and complete loss of chitin-induced calcium spiking in the Lyk5-RNAi background. The positive selection nature of APTi represents a fundamental improvement in RNAi technology and will contribute to the growing demand for technologies amenable to high-throughput phenotyping.
中文翻译:

用于腺嘌呤磷酸核糖基转移酶串联沉默的基因家族的基于生存的稳健 RNAi。
与 CRISPR-Cas 技术不同,RNA 干扰 (RNAi) 能够灵活动态地询问整个基因家族或必需基因,而不需要外源蛋白质。不幸的是,分离经历有效基因沉默的植物需要费力的设计、目视筛选和物理分离以进行下游表征。在这里,我们在小立碗藓中开发了一种基于腺嘌呤磷酸核糖基转移酶 (APT) 的 RNAi 技术 (APTi),该技术改进了当前 RNAi 技术的多重局限性。 APTi 利用 2-氟腺嘌呤存在下短暂沉默APT的促生存输出,从而将生存本身确立为 RNAi 的报告者。为了最大限度地提高基因靶标的沉默效果,我们创建了载体,以便于将任何基因靶标序列与 APT 沉默基序串联插入。我们测试了 APTi 对两个基因家族的功效,即肌动蛋白依赖性运动肌球蛋白 XI (a,b) 和假定的几丁质受体 Lyk5 (a,b,c)。 APTi 方法产生了对我们的基因靶标具有特异性的瞬态P. patens突变体的同质群体,在 8 天内存活的背景植物为零。观察到的突变体直接对应于 Lyk5-RNAi 背景中肌球蛋白 XI 蛋白最大减少 93% 和几丁质诱导的钙尖峰完全丧失。 APTi 的正选择性质代表了 RNAi 技术的根本性改进,并将有助于满足高通量表型分析技术不断增长的需求。











































 京公网安备 11010802027423号
京公网安备 11010802027423号