Structure ( IF 4.4 ) Pub Date : 2020-08-06 , DOI: 10.1016/j.str.2020.07.010 Zhou Gong 1 , Shang-Xiang Ye 2 , Chun Tang 3
|
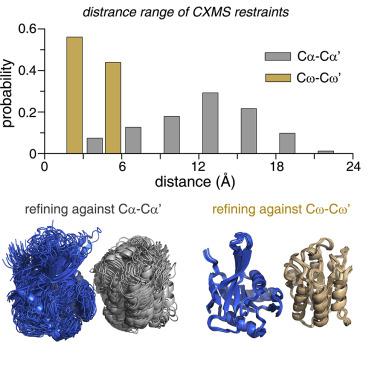
Chemical crosslinking coupled with mass spectrometry (CXMS) has been increasingly used in structural biology. CXMS distance restraints are usually applied to Cα or Cβ atoms of the crosslinked residues, with upper bounds typically over 20 Å. The incorporation of loose CXMS restraints only marginally improves the resolution of the calculated structures. Here, we present a revised format of CXMS distance restraints, which works by first modifying the crosslinked residue with a rigid extension derived from the crosslinker. With the flexible side chain explicitly represented, the reformatted restraint can be applied to the modification group instead, with an upper bound of 6 Å or less. The short distance restraint can be represented and back-calculated simply with a straight line. The use of tighter restraints not only afford better-resolved structures but also uncover protein dynamics. Together, our approach enables more information extracted from the CXMS data.
中文翻译:

收紧交联距离限制以更好地解析蛋白质结构和动力学。
化学交联与质谱联用 (CXMS) 已越来越多地用于结构生物学。CXMS 距离限制通常应用于交联残基的 Cα 或 Cβ 原子,上限通常超过 20 Å。松散 CXMS 约束的结合仅略微提高了计算结构的分辨率。在这里,我们提出了 CXMS 距离限制的修订格式,它的工作原理是首先用来自交联剂的刚性延伸修改交联残基。通过明确表示灵活的侧链,重新格式化的约束可以改为应用于修饰组,上限为 6 Å 或更小。短距离约束可以简单地用直线表示和反算。使用更严格的限制不仅可以提供更好的解析结构,还可以揭示蛋白质动力学。总之,我们的方法可以从 CXMS 数据中提取更多信息。









































 京公网安备 11010802027423号
京公网安备 11010802027423号