Acta Pharmaceutica Sinica B ( IF 14.7 ) Pub Date : 2020-08-05 , DOI: 10.1016/j.apsb.2020.07.020 Qian Wei , Jian Bai , Daojiang Yan , Xiuqi Bao , Wenting Li , Bingyu Liu , Dan Zhang , Xiangbing Qi , Dequan Yu , Youcai Hu
|
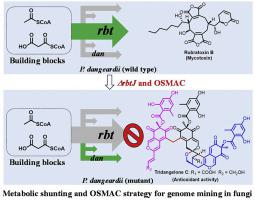
Endophytic fungi are promising producers of bioactive small molecules. Bioinformatic analysis of the genome of an endophytic fungus Penicillium dangeardii revealed 43 biosynthetic gene clusters, exhibited its strong ability to produce numbers of secondary metabolites. However, this strain mainly produce rubratoxins alone with high yield in varied culture conditions, suggested most gene clusters are silent. Efforts for mining the cryptic gene clusters in P. dangeardii, including epigenetic regulation and one-strain-many-compounds (OSMAC) approach were failed probably due to the high yield of rubratoxins. A metabolic shunting strategy by deleting the key gene for rubratoxins biosynthesis combining with optimization of culture condition successfully activated multiple silent genes encoding for other polyketide synthases (PKSs), and led to the trace compounds detectable. As a result, a total of 23 new compounds including azaphilone monomers, dimers, trimers with unprecedented polycyclic bridged heterocycle and spiral structures, as well as siderophores were identified. Some compounds showed significant cytotoxicities, anti-inflammatory or antioxidant activities. The attractive dual PKSs gene clusters for azaphilones biosynthesis were mined by bioinformatic analysis and overexpression of a pathway specific transcription factor. Our work therefor provides an efficient approach to mine the chemical diversity of endophytic fungi.
中文翻译:

基因组挖掘结合代谢分流和内生真菌的OSMAC策略导致产生多种天然产物
内生真菌是具有生物活性的小分子的有前途的生产者。对内生真菌丹氏青霉基因组的生物信息学分析揭示了43个生物合成基因簇,显示出其产生大量次生代谢产物的强大能力。然而,该菌株主要在不同的培养条件下仅以高产率产生雷巴托星,这表明大多数基因簇是沉默的。挖掘P. dangeardii隐性基因簇的努力包括表观遗传调控和一株多株化合物(OSMAC)方法的研究失败,可能是由于红曲霉毒素的高收率。通过删除用于维甲酸的生物合成关键基因并优化培养条件的代谢分流策略,成功激活了编码其他聚酮化合物合酶(PKS)的多个沉默基因,并导致可检测到痕量化合物。结果,鉴定出总共23种新化合物,包括氮杂苯甲酮单体,二聚体,具有空前多环桥联杂环和螺旋结构的三聚体以及铁载体。一些化合物显示出明显的细胞毒性,抗炎或抗氧化活性。通过生物信息学分析和途径特异性转录因子的过表达,挖掘出用于氮杂苯醌生物合成的有吸引力的双重PKSs基因簇。为此,我们的工作提供了一种有效的方法来挖掘内生真菌的化学多样性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号