Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2020-08-04 , DOI: 10.1016/j.jmgm.2020.107702 Ahmed H Al Sharie 1 , Tamam El-Elimat 2 , Yazan O Al Zu'bi 1 , Abdelwahab J Aleshawi 3 , José L Medina-Franco 4
|
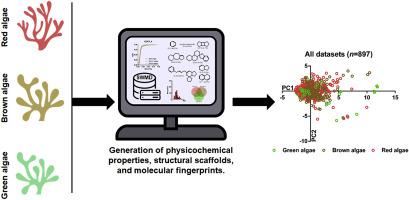
Seaweeds have attracted attention in the past decade as a biological source of highly diverse secondary metabolites with great potential in the industrial and pharmaceutical sciences. Herein, we represent a comprehensive cheminformatics study to compare the chemical diversity of seaweed metabolites based on their taxonomic source. Seaweed Metabolite Database (SWMD) was utilized in this study. The compounds were manually categorized into three datasets, namely red algae (Rhodophyta, n = 645), brown algae (Phaeophyta, n = 220), and green algae (Chlorophyta, n = 32). The compounds in each dataset were curated to generate six chemical descriptors of pharmaceutical interest for each molecule, which were later used to visualize the chemical space of these metabolites by principal component analysis. Scaffolds were generated by removing side chains and keeping the core part of each molecule. Scaffold diversity among the tested datasets was quantified using Cyclic System Retrieval Curves. Green algae metabolites in SWMD possessed the highest scaffold diversity followed by brown and red algae metabolites, respectively. Three structural binary fingerprints, including ECFP_4, MACCS keys, and PubChem were computed indicating that the red algae metabolites had the highest fingerprint diversity followed by the green and brown algae metabolites respectively. Finally, Consensus Diversity Plots were generated to assess the global diversity considering both scaffold and fingerprint diversity. It was concluded that green algae metabolites in the SWMD are the most diverse regarding chemical descriptors of pharmaceutical relevance and scaffolds. While red algae possess the highest fingerprint diversity.
中文翻译:

海藻代谢物数据库(SWMD)的化学空间和多样性:化学信息学研究。
在过去的十年中,海藻作为高度多样化的次生代谢物的生物来源而引起了人们的关注,在工业和制药科学中具有巨大的潜力。在此,我们进行了一项全面的化学信息学研究,根据海藻代谢物的分类来源来比较其化学多样性。本研究使用了海藻代谢物数据库(SWMD)。这些化合物被手动分类为三个数据集,即红藻(Rhodophyta,n = 645)、褐藻(Phaeophyta,n = 220)和绿藻(Chlorophyta,n = 32)。每个数据集中的化合物都经过精心设计,为每个分子生成六种具有制药意义的化学描述符,随后通过主成分分析将这些代谢物的化学空间可视化。通过去除侧链并保留每个分子的核心部分来生成支架。使用循环系统检索曲线对测试数据集之间的支架多样性进行量化。 SWMD 中的绿藻代谢物具有最高的支架多样性,其次是褐藻和红藻代谢物。计算了三个结构二元指纹,包括 ECFP_4、MACCS 键和 PubChem,表明红藻代谢物具有最高的指纹多样性,其次是绿藻和褐藻代谢物。最后,生成共识多样性图来评估考虑支架和指纹多样性的全局多样性。结论是,SWMD 中的绿藻代谢物在药物相关性和支架的化学描述符方面是最多样化的。而红藻则拥有最高的指纹多样性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号