Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A functional m6A‐RNA methylation pathway in the oyster Crassostrea gigas assumes epitranscriptomic regulation of lophotrochozoan development
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-08-02 , DOI: 10.1111/febs.15500 Lorane Le Franc 1, 2 , Benoit Bernay 3 , Bruno Petton 4 , Marc Since 5 , Pascal Favrel 1, 2 , Guillaume Rivière 1, 2
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-08-02 , DOI: 10.1111/febs.15500 Lorane Le Franc 1, 2 , Benoit Bernay 3 , Bruno Petton 4 , Marc Since 5 , Pascal Favrel 1, 2 , Guillaume Rivière 1, 2
Affiliation
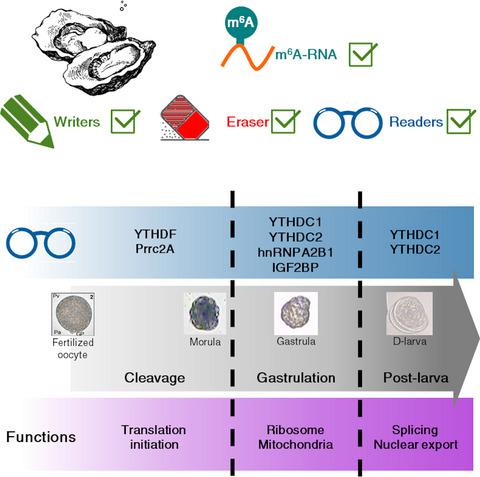
|
N6‐methyladenosine (m6A) is a prevalent epitranscriptomic mark in eukaryotic RNA, with crucial roles for mammalian and ecdysozoan development. Indeed, m6A‐RNA and the related protein machinery are important for splicing, translation, maternal‐to‐zygotic transition and cell differentiation. However, to date, the presence of an m6A‐RNA pathway remains unknown in more distant animals, questioning the evolution and significance of the epitranscriptomic regulation. Therefore, we investigated the m6A‐RNA pathway in the oyster Crassostrea gigas, a lophotrochozoan model whose development was demonstrated under strong epigenetic influence. Using mass spectrometry and dot blot assays, we demonstrated that m6A‐RNA is actually present in the oyster and displays variations throughout early oyster development, with the lowest levels at the end of cleavage. In parallel, by in silico analyses, we were able to characterize at the molecular level a complete and conserved putative m6A machinery. The expression levels of the identified putative m6A writers, erasers and readers were strongly regulated across oyster development. Finally, RNA pull‐down coupled to LC‐MS/MS allowed us to prove the actual presence of readers able to bind m6A‐RNA and exhibiting specific developmental patterns. Altogether, our results demonstrate the conservation of a complete m6A‐RNA pathway in the oyster and strongly suggest its implication in early developmental processes including MZT. This first demonstration and characterization of an epitranscriptomic regulation in a lophotrochozoan model, potentially involved in the embryogenesis, bring new insights into our understanding of developmental epigenetic processes and their evolution.
中文翻译:

牡蛎Crassostrea gigas 中的功能性m6A-RNA 甲基化途径假定表观转录组学调控原虫发育
N 6甲基腺苷(m 6 A) 是真核生物 RNA 中普遍存在的表观转录组标记,对哺乳动物和蜕皮动物的发育起着至关重要的作用。事实上,m 6 A-RNA 和相关的蛋白质机制对于剪接、翻译、母体到合子的转变和细胞分化很重要。然而,迄今为止,m 6 A-RNA 通路在更远距离的动物中的存在仍然未知,这对表观转录组调控的进化和重要性提出了质疑。因此,我们研究了牡蛎Crassostrea gigas 中的 m 6 A-RNA 通路。,一种 lohotrochozoan 模型,其发展在强烈的表观遗传影响下得到证明。使用质谱法和斑点印迹分析,我们证明 m 6 A-RNA 实际上存在于牡蛎中,并且在牡蛎早期发育过程中表现出变化,在卵裂结束时水平最低。同时,通过计算机分析,我们能够在分子水平上表征完整且保守的推定 m 6 A 机制。确定的推定 m 6 A 书写器、橡皮擦和阅读器的表达水平在牡蛎发育过程中受到强烈调节。最后,结合 LC-MS/MS 的 RNA pull-down 使我们能够证明能够结合 m 6的阅读器的实际存在A-RNA 并表现出特定的发育模式。总而言之,我们的结果证明了牡蛎中完整的 m 6 A-RNA 通路的保守性,并强烈表明其对包括 MZT 在内的早期发育过程的影响。首次证明和表征了可能参与胚胎发生的 lophotrochozoan 模型中的表观转录组调控,为我们对发育表观遗传过程及其进化的理解带来了新的见解。
更新日期:2020-08-02
中文翻译:

牡蛎Crassostrea gigas 中的功能性m6A-RNA 甲基化途径假定表观转录组学调控原虫发育
N 6甲基腺苷(m 6 A) 是真核生物 RNA 中普遍存在的表观转录组标记,对哺乳动物和蜕皮动物的发育起着至关重要的作用。事实上,m 6 A-RNA 和相关的蛋白质机制对于剪接、翻译、母体到合子的转变和细胞分化很重要。然而,迄今为止,m 6 A-RNA 通路在更远距离的动物中的存在仍然未知,这对表观转录组调控的进化和重要性提出了质疑。因此,我们研究了牡蛎Crassostrea gigas 中的 m 6 A-RNA 通路。,一种 lohotrochozoan 模型,其发展在强烈的表观遗传影响下得到证明。使用质谱法和斑点印迹分析,我们证明 m 6 A-RNA 实际上存在于牡蛎中,并且在牡蛎早期发育过程中表现出变化,在卵裂结束时水平最低。同时,通过计算机分析,我们能够在分子水平上表征完整且保守的推定 m 6 A 机制。确定的推定 m 6 A 书写器、橡皮擦和阅读器的表达水平在牡蛎发育过程中受到强烈调节。最后,结合 LC-MS/MS 的 RNA pull-down 使我们能够证明能够结合 m 6的阅读器的实际存在A-RNA 并表现出特定的发育模式。总而言之,我们的结果证明了牡蛎中完整的 m 6 A-RNA 通路的保守性,并强烈表明其对包括 MZT 在内的早期发育过程的影响。首次证明和表征了可能参与胚胎发生的 lophotrochozoan 模型中的表观转录组调控,为我们对发育表观遗传过程及其进化的理解带来了新的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号