当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Slice profile effects on quantitative analysis of hyperpolarized pyruvate.
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-08-02 , DOI: 10.1002/nbm.4373 Christopher M Walker 1 , Jeremy W Gordon 2 , Zhan Xu 1 , Keith A Michel 1, 3 , Liang Li 4 , Peder E Z Larson 2 , Daniel B Vigneron 2 , James A Bankson 1, 3
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-08-02 , DOI: 10.1002/nbm.4373 Christopher M Walker 1 , Jeremy W Gordon 2 , Zhan Xu 1 , Keith A Michel 1, 3 , Liang Li 4 , Peder E Z Larson 2 , Daniel B Vigneron 2 , James A Bankson 1, 3
Affiliation
|
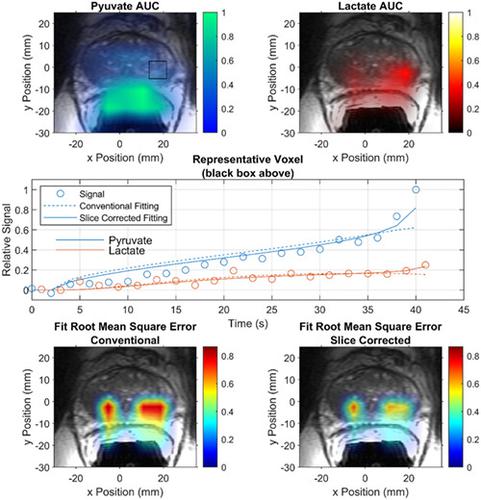
Magnetic resonance imaging of hyperpolarized pyruvate provides a new imaging biomarker for cancer metabolism, based on the dynamic in vivo conversion of hyperpolarized pyruvate to lactate. Methods for quantification of signal evolution need to be robust and reproducible across a range of experimental conditions. Pharmacokinetic analysis of dynamic spectroscopic imaging data from hyperpolarized pyruvate and its metabolites generally assumes that signal arises from ideal rectangular slice excitation profiles. In this study, we examined whether this assumption could lead to bias in kinetic analysis of hyperpolarized pyruvate and, if so, whether such a bias can be corrected. A Bloch‐McConnell simulator was used to generate synthetic data using a known set of “ground truth” pharmacokinetic parameter values. Signal evolution was then analyzed using analysis software that either assumed a uniform slice profile, or incorporated information about the slice profile into the analysis. To correct for slice profile effects, the expected slice profile was subdivided into multiple sub‐slices to account for variable excitation angles along the slice dimension. An ensemble of sub‐slices was then used to fit the measured signal evolution. A mismatch between slice profiles used for data acquisition and those assumed during kinetic analysis was identified as a source of quantification bias. Results indicate that imperfect slice profiles preferentially increase detected lactate signal, leading to an overestimation of the apparent metabolic exchange rate. The slice profile‐correction algorithm was tested in simulation, in phantom measurements, and applied to data acquired from a patient with prostate cancer. The results demonstrated that slice profile‐induced biases can be minimized by accounting for the slice profile during pharmacokinetic analysis. This algorithm can be used to correct data from either single or multislice acquisitions.
中文翻译:

切片剖面对超极化丙酮酸定量分析的影响。
基于超极化丙酮酸在体内动态转化为乳酸,超极化丙酮酸的磁共振成像为癌症代谢提供了一种新的成像生物标志物。信号演变的量化方法需要在一系列实验条件下稳健且可重现。来自超极化丙酮酸及其代谢物的动态光谱成像数据的药代动力学分析通常假设信号来自理想的矩形切片激发剖面。在这项研究中,我们检查了这种假设是否会导致超极化丙酮酸动力学分析的偏差,如果是,这种偏差是否可以得到纠正。Bloch-McConnell 模拟器用于使用一组已知的“真实”药代动力学参数值生成合成数据。然后使用分析软件对信号演变进行分析,该软件假设一个统一的切片剖面,或者将有关切片剖面的信息纳入分析。为了校正切片剖面效应,预期切片剖面被细分为多个子切片,以考虑沿切片尺寸的可变激发角。然后使用一组子切片来拟合测量的信号演变。用于数据采集的切片轮廓与动力学分析期间假设的切片轮廓之间的不匹配被确定为量化偏差的来源。结果表明,不完美的切片配置文件优先增加检测到的乳酸信号,导致表观代谢交换率的高估。切片轮廓校正算法在模拟中进行了测试,在体模测量中,并应用于从前列腺癌患者那里获得的数据。结果表明,通过在药代动力学分析过程中考虑切片剖面,可以最大限度地减少切片剖面引起的偏差。该算法可用于校正来自单切片或多切片采集的数据。
更新日期:2020-09-03
中文翻译:

切片剖面对超极化丙酮酸定量分析的影响。
基于超极化丙酮酸在体内动态转化为乳酸,超极化丙酮酸的磁共振成像为癌症代谢提供了一种新的成像生物标志物。信号演变的量化方法需要在一系列实验条件下稳健且可重现。来自超极化丙酮酸及其代谢物的动态光谱成像数据的药代动力学分析通常假设信号来自理想的矩形切片激发剖面。在这项研究中,我们检查了这种假设是否会导致超极化丙酮酸动力学分析的偏差,如果是,这种偏差是否可以得到纠正。Bloch-McConnell 模拟器用于使用一组已知的“真实”药代动力学参数值生成合成数据。然后使用分析软件对信号演变进行分析,该软件假设一个统一的切片剖面,或者将有关切片剖面的信息纳入分析。为了校正切片剖面效应,预期切片剖面被细分为多个子切片,以考虑沿切片尺寸的可变激发角。然后使用一组子切片来拟合测量的信号演变。用于数据采集的切片轮廓与动力学分析期间假设的切片轮廓之间的不匹配被确定为量化偏差的来源。结果表明,不完美的切片配置文件优先增加检测到的乳酸信号,导致表观代谢交换率的高估。切片轮廓校正算法在模拟中进行了测试,在体模测量中,并应用于从前列腺癌患者那里获得的数据。结果表明,通过在药代动力学分析过程中考虑切片剖面,可以最大限度地减少切片剖面引起的偏差。该算法可用于校正来自单切片或多切片采集的数据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号