Letters in Drug Design & Discovery ( IF 1.2 ) Pub Date : 2020-07-31 , DOI: 10.2174/1570180817666200108095912 Reaz Uddin 1 , Bushra Siraj 1 , Sidra Rafi 1 , Syed Sikander Azam 2 , Abdul Wadood 3
|
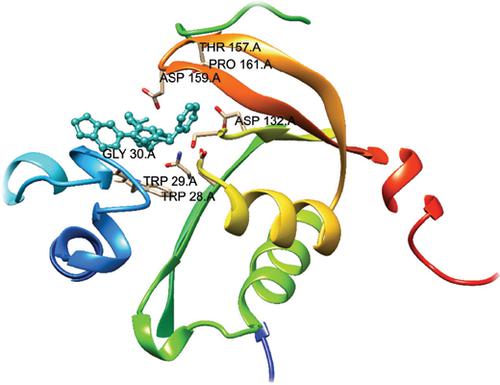
Background: Aminoglycoside 6'-N-acetyltransferase type Ib (AAC(6')-Ib) from Klebsiella pneumoniae is an established drug target and has conferred insensitivity to aminoglycosides. Aminoglycosides are often inactivated by aminoglycoside modifying enzymes encoded by genes present in the chromosome, plasmids, and other genetic elements. The AAC(6′)- Ib is an enzyme of clinical importance found in a wide variety of gram-negative pathogens. The AAC(6′)-Ib enzyme is of interest not only because of its ubiquity but also because of other characteristics e.g., it presents significant microheterogeneity at the N-termini and the aac(6′)-Ib gene is often present in integrons, transposons, plasmids, genomic islands, and other genetic structures. The majority of the reported potent inhibitors against the target are substrate analogs. Therefore, there is a need to develop or discover new scaffolds other than substrate analogs as AAC(6')-Ib inhibitor.
Objective: The objective of this study is to set optimum parameters for the structure-based virtual screening by multiple docking and scoring methods. The multiple scoring of each ligand also incorporates the ‘Induced Fit’ docking effect that helps to build further confidence in the shortlisted compounds. The method eventually is able to predict the potential inhibitors that bind to the active site and can potentially inhibit the activity of the Aminoglycoside 6′-N-acetyltransferase type Ib [AAC(6’)-Ib] from Klebsiella pneumoniae.
Methods: Using the available three-dimensional structure of enzyme AAC(6')-Ib inhibitor complex, a structure-based virtual screening was performed with the hope of prioritizing the promising leads. In order to set up the protocol, 30,000 drug-like molecules were selected from the ChemBridge library. Multiple docking programs, i.e. UCSF DOCK6 and AutoDock Vina have been applied in the current study so that a consensus is developed to the predicted binding modes and thus the docking accuracy. The Amber scores of the Dock6 – a secondary scoring function was also used to perform the ‘Induced Fit’ effect and correspondingly re-rank the compounds.
Results: The top 30 ranked compounds of the most frequent scored were selected from the histogram. The 2D interactions of those 30 compounds were drawn from the Ligplot+ tool. Six of the compounds were prioritized as potential inhibitors as they are representing the maximum number of interactions from the rest of the compounds and also possess the drug-likeness as predicted by the estimated ADMET properties.
Conclusion: This study provided useful insight that the proposed compounds have the potential to bind to the aminoglycoside binding site of AAC(6′)-Ib that may eventually inhibit the Klebsiella pneumoniae. This study has the potential to propose putative new and novel inhibitors against a resistant drug target of Klebsiella pneumoniae.
中文翻译:

基于结构的虚拟筛选方法,发现针对肺炎克雷伯菌感染的氨基糖苷6'-N-乙酰基转移酶Ib型[AAC(6')-Ib]有效抑制剂
背景:来自肺炎克雷伯菌的氨基糖苷6'-N-乙酰基转移酶Ib型(AAC(6')-Ib)是既定的药物靶标,赋予了氨基糖苷类药物不敏感性。氨基糖苷通常被染色体,质粒和其他遗传元件中存在的基因编码的氨基糖苷修饰酶灭活。AAC(6')-Ib是在多种革兰氏阴性病原体中发现的具有临床重要性的酶。AAC(6')-Ib酶不仅因为其普遍存在,还因为其他特征而受到关注,例如,它在N-末端具有明显的微异质性,而aac(6')-Ib基因通常存在于整合素中,转座子,质粒,基因组岛和其他遗传结构。报道的大多数针对靶标的有效抑制剂是底物类似物。因此,
目的:本研究的目的是通过多种对接和评分方法为基于结构的虚拟筛选设置最佳参数。每个配体的多重得分还结合了“诱导契合”对接效应,有助于进一步入围化合物。该方法最终能够预测与活性位点结合的潜在抑制剂,并且可以潜在地抑制肺炎克雷伯菌的氨基糖苷6'-N-乙酰基转移酶Ib [AAC(6')-Ib]的活性。
方法:利用酶AAC(6')-Ib抑制剂复合物的可用三维结构,进行了基于结构的虚拟筛选,希望优先考虑有前途的潜在客户。为了建立协议,从ChemBridge库中选择了30,000种药物样分子。在当前的研究中已经应用了多个对接程序,即UCSF DOCK6和AutoDock Vina,以便对预测的绑定模式以及对接精度达成共识。Dock6的Amber评分-次要评分功能也用于执行“诱导契合”效果并相应地对化合物进行重新排名。
结果:从直方图中选择得分最高的前30位化合物。从Ligplot +工具中提取了这30种化合物的2D相互作用。其中六个化合物被优先考虑作为潜在的抑制剂,因为它们代表了其余化合物的最大相互作用数,并且还具有如估计的ADMET特性所预测的类似药物的特性。
结论:这项研究提供了有用的见解,认为拟议的化合物具有与AAC(6')-Ib的氨基糖苷结合位点结合的潜力,这可能最终抑制肺炎克雷伯菌。这项研究有可能提出针对肺炎克雷伯菌耐药药物靶点的新型抑制剂。











































 京公网安备 11010802027423号
京公网安备 11010802027423号