当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Naked Bacterium: Emerging Properties of a Surfome-Streamlined Pseudomonas putida Strain.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-07-30 , DOI: 10.1021/acssynbio.0c00272 Esteban Martínez-García 1 , Sofía Fraile 1 , David Rodríguez Espeso 1 , Davide Vecchietti 2 , Giovanni Bertoni 2 , Víctor de Lorenzo 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-07-30 , DOI: 10.1021/acssynbio.0c00272 Esteban Martínez-García 1 , Sofía Fraile 1 , David Rodríguez Espeso 1 , Davide Vecchietti 2 , Giovanni Bertoni 2 , Víctor de Lorenzo 1
Affiliation
|
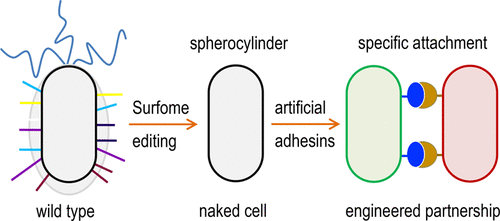
Environmental bacteria are most often endowed with native surface-attachment programs that frequently conflict with efforts to engineer biofilms and synthetic communities with given tridimensional architectures. In this work, we report the editing of the genome of Pseudomonas putida KT2440 for stripping the cells of most outer-facing structures of the bacterial envelope that mediate motion, binding to surfaces, and biofilm formation. To this end, 23 segments of the P. putida chromosome encoding a suite of such functions were deleted, resulting in the surface-naked strain EM371, the physical properties of which changed dramatically in respect to the wild type counterpart. As a consequence, surface-edited P. putida cells were unable to form biofilms on solid supports and, because of the swimming deficiency and other alterations, showed a much faster sedimentation in liquid media. Surface-naked bacteria were then used as carriers of interacting partners (e.g., Jun–Fos domains) ectopically expressed by means of an autotransporter display system on the now easily accessible cell envelope. Abstraction of individual bacteria as adhesin-coated spherocylinders enabled rigorous quantitative description of the multicell interplay brought about by thereby engineered physical interactions. The model was then applied to parametrize the data extracted from automated analysis of confocal microscopy images of the experimentally assembled bacterial flocks for analyzing their structure and distribution. The resulting data not only corroborated the value of P. putida EM371 over the parental strain as a platform for display artificial adhesins but also provided a strategy for rational engineering of catalytic communities.
中文翻译:

裸细菌: Surfome-Streamlined Pseudomonas putida 菌株的新兴特性。
环境细菌通常具有天然的表面附着程序,这些程序经常与设计具有给定三维结构的生物膜和合成群落的努力相冲突。在这项工作中,我们报告了恶臭假单胞菌KT2440基因组的编辑,用于剥离细菌包膜的大多数外向结构的细胞,这些结构介导运动、与表面结合和生物膜形成。为此,编码一组此类功能的恶臭假单胞菌染色体的23 个片段被删除,导致表面裸体菌株 EM371,其物理特性相对于野生型对应物发生了巨大变化。因此,表面编辑的恶臭假单胞菌细胞无法在固体支持物上形成生物膜,并且由于游泳不足和其他改变,在液体介质中的沉降速度要快得多。然后将表面裸露的细菌用作相互作用伙伴(例如 Jun-Fos 结构域)的载体,通过现在易于访问的细胞包膜上的自动转运蛋白展示系统异位表达。将单个细菌抽象为粘附素涂层的圆柱体,可以对由此工程物理相互作用带来的多细胞相互作用进行严格的定量描述。然后应用该模型对从实验组装的细菌群的共聚焦显微镜图像的自动分析中提取的数据进行参数化,以分析它们的结构和分布。结果数据不仅证实了P. putida EM371 在亲本菌株上作为展示人工粘附素的平台,但也为催化群落的合理工程提供了策略。
更新日期:2020-09-20
中文翻译:

裸细菌: Surfome-Streamlined Pseudomonas putida 菌株的新兴特性。
环境细菌通常具有天然的表面附着程序,这些程序经常与设计具有给定三维结构的生物膜和合成群落的努力相冲突。在这项工作中,我们报告了恶臭假单胞菌KT2440基因组的编辑,用于剥离细菌包膜的大多数外向结构的细胞,这些结构介导运动、与表面结合和生物膜形成。为此,编码一组此类功能的恶臭假单胞菌染色体的23 个片段被删除,导致表面裸体菌株 EM371,其物理特性相对于野生型对应物发生了巨大变化。因此,表面编辑的恶臭假单胞菌细胞无法在固体支持物上形成生物膜,并且由于游泳不足和其他改变,在液体介质中的沉降速度要快得多。然后将表面裸露的细菌用作相互作用伙伴(例如 Jun-Fos 结构域)的载体,通过现在易于访问的细胞包膜上的自动转运蛋白展示系统异位表达。将单个细菌抽象为粘附素涂层的圆柱体,可以对由此工程物理相互作用带来的多细胞相互作用进行严格的定量描述。然后应用该模型对从实验组装的细菌群的共聚焦显微镜图像的自动分析中提取的数据进行参数化,以分析它们的结构和分布。结果数据不仅证实了P. putida EM371 在亲本菌株上作为展示人工粘附素的平台,但也为催化群落的合理工程提供了策略。











































 京公网安备 11010802027423号
京公网安备 11010802027423号