当前位置:
X-MOL 学术
›
J. Syst. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Resolving robust phylogenetic relationships of core Brassicaceae using genome skimming data
Journal of Systematics and Evolution ( IF 3.7 ) Pub Date : 2020-07-29 , DOI: 10.1111/jse.12666 Liang‐Min Liu 1, 2 , Xin‐Yu Du 1, 2 , Cen Guo 1 , De‐Zhu Li 1, 2
Journal of Systematics and Evolution ( IF 3.7 ) Pub Date : 2020-07-29 , DOI: 10.1111/jse.12666 Liang‐Min Liu 1, 2 , Xin‐Yu Du 1, 2 , Cen Guo 1 , De‐Zhu Li 1, 2
Affiliation
|
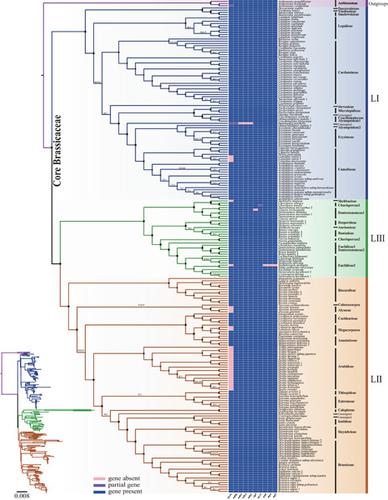
The Brassicaceae is an economically and scientifically important family distributed globally, including oilseed rape and the model plant, Arabidopsis thaliana. Although growing molecular data have been used in phylogenetic studies, the relationships among major clades and tribes of Brassicaceae are still controversial. Here, we investigated the core Brassicaceae phylogenetics using 222 plastomes and 235 nrDNA cistrons, including 106 plastomes and 112 nrDNA cistrons assembled from newly sequenced genome skimming data of 112 taxa. The sampling covered 73 genera from 61.5% tribes and four unassigned genera and species. Three well supported lineages LI, LII, and LIII were revealed in our plastomic analyses, with LI sister to LII + LIII. In addition, the monophyly of the newly delimitated LII was strongly supported by three different partition strategies, concatenated methods under Bayesian and Maximum Likelihood analyses. LII comprised 13 tribes, including four tribes previously unassigned to any lineage, that is Biscutelleae as the earliest diverging clade and Cochlearieae as the sister to Megacarpaeeae + Anastaticeae. Within LII, the intertribal relationships were also well resolved, except that a conflicting position of Orychophragmus was detected among different datasets. In LIII, Shehbazia was resolved as a member of Chorisproreae, but Chorisproreae, Dontostemoneae, and Euclidieae were all resolved as paraphyletic, which was also confirmed by nrDNA analyses. Moreover, the loss of the rps16 gene was detected as likely to be a synapomorphy of the tribes Arabideae and Alysseae. Overall, using genome skimming data, we resolved robust phylogenetic relationships of core Brassicaceae and shed new light on the complex evolutionary history of this family.
中文翻译:

使用基因组略读数据解析核心十字花科的强大系统发育关系
十字花科是一个在经济和科学上分布于全球的重要科,包括油菜和模式植物拟南芥. 尽管在系统发育研究中使用了越来越多的分子数据,但十字花科主要进化枝和部落之间的关系仍然存在争议。在这里,我们使用 222 个塑性体和 235 个 nrDNA 顺反子研究了十字花科的核心系统发育,其中包括从 112 个分类群的新测序的基因组撇取数据组装的 106 个塑性体和 112 个 nrDNA 顺反子。抽样涵盖了来自 61.5% 部落的 73 个属和四个未指定的属和种。在我们的塑性体分析中揭示了三个得到良好支持的谱系 LI、LII 和 LIII,其中 LI 是 LII + LIII 的姐妹。此外,新界定的 LII 的单一性得到了三种不同的分区策略、贝叶斯和最大似然分析下的连接方法的强烈支持。LII 由 13 个部落组成,其中包括 4 个以前未分配给任何血统的部落,即 Biscutelleae 作为最早的分支进化枝和 Cochlearieae 作为 Megacarpaeeae + Anastaticeae 的姐妹。在 LII 内部,部落间关系也得到了很好的解决,除了在不同的数据集中检测到Orychophragmus。在 LIII 中,Shehbazia被解析为Chorisproreae的成员,但 Chorisproreae、Dontostemoneae 和 Euclidieae 都被解析为并系群,这也得到了 nrDNA 分析的证实。此外,检测到rps16基因的丢失可能是阿拉伯科和 Alysseae 部落的突触。总体而言,使用基因组浏览数据,我们解决了核心十字花科的强大系统发育关系,并为该科复杂的进化历史提供了新的线索。
更新日期:2020-07-29
中文翻译:

使用基因组略读数据解析核心十字花科的强大系统发育关系
十字花科是一个在经济和科学上分布于全球的重要科,包括油菜和模式植物拟南芥. 尽管在系统发育研究中使用了越来越多的分子数据,但十字花科主要进化枝和部落之间的关系仍然存在争议。在这里,我们使用 222 个塑性体和 235 个 nrDNA 顺反子研究了十字花科的核心系统发育,其中包括从 112 个分类群的新测序的基因组撇取数据组装的 106 个塑性体和 112 个 nrDNA 顺反子。抽样涵盖了来自 61.5% 部落的 73 个属和四个未指定的属和种。在我们的塑性体分析中揭示了三个得到良好支持的谱系 LI、LII 和 LIII,其中 LI 是 LII + LIII 的姐妹。此外,新界定的 LII 的单一性得到了三种不同的分区策略、贝叶斯和最大似然分析下的连接方法的强烈支持。LII 由 13 个部落组成,其中包括 4 个以前未分配给任何血统的部落,即 Biscutelleae 作为最早的分支进化枝和 Cochlearieae 作为 Megacarpaeeae + Anastaticeae 的姐妹。在 LII 内部,部落间关系也得到了很好的解决,除了在不同的数据集中检测到Orychophragmus。在 LIII 中,Shehbazia被解析为Chorisproreae的成员,但 Chorisproreae、Dontostemoneae 和 Euclidieae 都被解析为并系群,这也得到了 nrDNA 分析的证实。此外,检测到rps16基因的丢失可能是阿拉伯科和 Alysseae 部落的突触。总体而言,使用基因组浏览数据,我们解决了核心十字花科的强大系统发育关系,并为该科复杂的进化历史提供了新的线索。



























 京公网安备 11010802027423号
京公网安备 11010802027423号