Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Searching for remote homologs of CAML among eukaryotes.
Traffic ( IF 3.6 ) Pub Date : 2020-07-26 , DOI: 10.1111/tra.12758 Nica Borgese 1
Traffic ( IF 3.6 ) Pub Date : 2020-07-26 , DOI: 10.1111/tra.12758 Nica Borgese 1
Affiliation
|
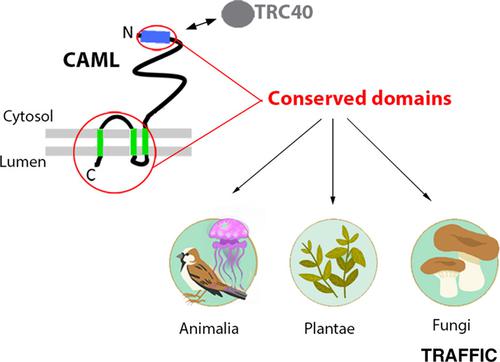
The tryptophan rich basic protein/calcium signal‐modulating cyclophilin ligand (WRB/CAML) and Get1p/Get2p complexes, in vertebrates and yeast, respectively, mediate the final step of tail‐anchored protein insertion into the endoplasmic reticulum membrane via the Get pathway. While WRB appears to exist in all eukaryotes, CAML homologs were previously recognized only among chordates, raising the question as to how CAML's function is performed in other phyla. Furthermore, whereas WRB was recognized as the metazoan homolog of Get1, CAML and Get2, although functionally equivalent, were not considered to be homologous. CAML contains an N‐terminal basic, TRC40/Get3‐interacting, region, three transmembrane segments near the C‐terminus, and a poorly conserved region between these domains. Here, I searched the NCBI protein database for remote CAML homologs in all eukaryotes, using position‐specific iterated‐basic local alignment search tool, with the C‐terminal, the N‐terminal or the full‐length sequence of human CAML as query. The N‐terminal basic region and full‐length CAML retrieved homologs among metazoa, plants and fungi. In the latter group several hits were annotated as GET2. The C‐terminal query did not return entries outside of the animal kingdom, but did retrieve over one hundred invertebrate metazoan CAML‐like proteins, which all conserved the N‐terminal TRC40‐binding domain. The results indicate that CAML homologs exist throughout the eukaryotic domain of life, and suggest that metazoan CAML and yeast GET2 share a common evolutionary origin. They further reveal a tight link between the particular features of the metazoan membrane‐anchoring domain and the TRC40‐interacting region. The list of sequences presented here should provide a useful resource for future studies addressing structure‐function relationships in CAML proteins.
中文翻译:

在真核生物中搜索CAML的远程同源物。
分别在脊椎动物和酵母中富含色氨酸的碱性蛋白/钙信号调节亲环蛋白配体(WRB / CAML)和Get1p / Get2p复合物介导通过Get途径将尾锚蛋白插入内质网的最后一步。虽然WRB似乎在所有真核生物中都存在,但以前仅在脊索动物中才认识到CAML同源物,这引发了一个问题,即CAML在其他门中如何发挥作用。此外,虽然WRB被认为是Get1的后生同源物,但CAML和Get2尽管功能上等效,但并不被认为是同源的。CAML包含一个N末端基本,TRC40 / Get3相互作用区域,C末端附近的三个跨膜片段,以及这些域之间的保守性较差的区域。这里,我使用位置特定的迭代基础局部比对搜索工具在人类CAML的C末端,N末端或全长序列中进行查询,从而在所有真核生物中的NCBI蛋白质数据库中搜索了远程CAML同源物。N末端基本区域和全长CAML检索到后生动物,植物和真菌之间的同源物。在后一组中,一些命中标记为GET2。C末端查询未返回动物界以外的条目,但确实检索了一百多种无脊椎动物后生动物CAML样蛋白,它们均保留了N末端TRC40结合域。结果表明,CAML同源物存在于整个生命的真核域,并表明后生动物的CAML和酵母GET2具有共同的进化起源。他们进一步揭示了后生膜锚定域的特殊特征与TRC40相互作用区域之间的紧密联系。此处列出的序列清单应为将来研究CAML蛋白中的结构功能关系提供有用的资源。
更新日期:2020-09-25
中文翻译:

在真核生物中搜索CAML的远程同源物。
分别在脊椎动物和酵母中富含色氨酸的碱性蛋白/钙信号调节亲环蛋白配体(WRB / CAML)和Get1p / Get2p复合物介导通过Get途径将尾锚蛋白插入内质网的最后一步。虽然WRB似乎在所有真核生物中都存在,但以前仅在脊索动物中才认识到CAML同源物,这引发了一个问题,即CAML在其他门中如何发挥作用。此外,虽然WRB被认为是Get1的后生同源物,但CAML和Get2尽管功能上等效,但并不被认为是同源的。CAML包含一个N末端基本,TRC40 / Get3相互作用区域,C末端附近的三个跨膜片段,以及这些域之间的保守性较差的区域。这里,我使用位置特定的迭代基础局部比对搜索工具在人类CAML的C末端,N末端或全长序列中进行查询,从而在所有真核生物中的NCBI蛋白质数据库中搜索了远程CAML同源物。N末端基本区域和全长CAML检索到后生动物,植物和真菌之间的同源物。在后一组中,一些命中标记为GET2。C末端查询未返回动物界以外的条目,但确实检索了一百多种无脊椎动物后生动物CAML样蛋白,它们均保留了N末端TRC40结合域。结果表明,CAML同源物存在于整个生命的真核域,并表明后生动物的CAML和酵母GET2具有共同的进化起源。他们进一步揭示了后生膜锚定域的特殊特征与TRC40相互作用区域之间的紧密联系。此处列出的序列清单应为将来研究CAML蛋白中的结构功能关系提供有用的资源。











































 京公网安备 11010802027423号
京公网安备 11010802027423号