当前位置:
X-MOL 学术
›
Microb. Biotechnol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
SEVA 3.1: enabling interoperability of DNA assembly among the SEVA, BioBricks and Type IIS restriction enzyme standards.
Microbial Biotechnology ( IF 4.8 ) Pub Date : 2020-07-25 , DOI: 10.1111/1751-7915.13609 Stamatios G Damalas 1 , Christos Batianis 1 , Maria Martin-Pascual 1 , Victor de Lorenzo 2 , Vitor A P Martins Dos Santos 1, 3
Microbial Biotechnology ( IF 4.8 ) Pub Date : 2020-07-25 , DOI: 10.1111/1751-7915.13609 Stamatios G Damalas 1 , Christos Batianis 1 , Maria Martin-Pascual 1 , Victor de Lorenzo 2 , Vitor A P Martins Dos Santos 1, 3
Affiliation
|
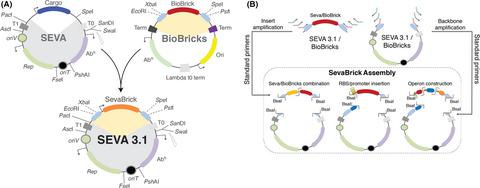
Robust synthetic biology applications rely heavily on the design and assembly of DNA parts with specific functionalities based on engineering principles. However, the assembly standards adopted by different communities vary considerably, thus limiting the interoperability of parts, vectors and methods. We hereby introduce the SEVA 3.1 platform consisting of the SEVA 3.1 vectors and the Golden Gate‐based ‘SevaBrick Assembly’. This platform enables the convergence of standard processes between the SEVA platform, the BioBricks and the Type IIs‐mediated DNA assemblies to reduce complexity and optimize compatibility between parts and methods. It features a wide library of cloning vectors along with a core set of standard SevaBrick primers that allow multipart assembly and exchange of short functional genetic elements (promoters, RBSs) with minimal cloning and design effort. As proof of concept, we constructed, among others, multiple sfGFP expression vectors under the control of eight RBSs, eight promoters and four origins of replication as well as an inducible four‐gene operon expressing the biosynthetic genes for the black pigment proviolacein. To demonstrate the interoperability of the SEVA 3.1 vectors, all constructs were characterized in both Pseudomonas putida and Escherichia coli. In summary, the SEVA 3.1 platform optimizes compatibility and modularity of inserts and backbones with a cost‐ and time‐friendly DNA assembly method, substantially expanding the toolbox for successful synthetic biology applications in Gram‐negative bacteria.
中文翻译:

SEVA 3.1:实现 SEVA、BioBricks 和 IIS 型限制性内切酶标准之间 DNA 组装的互操作性。
强大的合成生物学应用在很大程度上依赖于基于工程原理的具有特定功能的 DNA 部件的设计和组装。然而,不同社区采用的装配标准差异很大,从而限制了零件、向量和方法的互操作性。我们在此介绍由 SEVA 3.1 矢量和基于 Golden Gate 的“SevaBrick Assembly”组成的 SEVA 3.1 平台。该平台实现了 SEVA 平台、BioBricks 和 Type II 介导的 DNA 组装之间标准流程的融合,以降低复杂性并优化部件和方法之间的兼容性。它具有广泛的克隆载体库以及一组核心标准 SevaBrick 引物,允许以最少的克隆和设计工作进行多部分组装和短功能遗传元件(启动子、RBS)的交换。作为概念证明,我们在八个 RBS、八个启动子和四个复制起点的控制下构建了多个sfGFP表达载体,以及表达黑色素原紫草素生物合成基因的诱导型四基因操纵子。为了证明 SEVA 3.1 载体的互操作性,所有构建体都在恶臭假单胞菌和大肠杆菌中进行了表征。总之,SEVA 3.1 平台通过成本和时间友好的 DNA 组装方法优化了插入物和主链的兼容性和模块化,大大扩展了革兰氏阴性细菌中成功合成生物学应用的工具箱。
更新日期:2020-07-25
中文翻译:

SEVA 3.1:实现 SEVA、BioBricks 和 IIS 型限制性内切酶标准之间 DNA 组装的互操作性。
强大的合成生物学应用在很大程度上依赖于基于工程原理的具有特定功能的 DNA 部件的设计和组装。然而,不同社区采用的装配标准差异很大,从而限制了零件、向量和方法的互操作性。我们在此介绍由 SEVA 3.1 矢量和基于 Golden Gate 的“SevaBrick Assembly”组成的 SEVA 3.1 平台。该平台实现了 SEVA 平台、BioBricks 和 Type II 介导的 DNA 组装之间标准流程的融合,以降低复杂性并优化部件和方法之间的兼容性。它具有广泛的克隆载体库以及一组核心标准 SevaBrick 引物,允许以最少的克隆和设计工作进行多部分组装和短功能遗传元件(启动子、RBS)的交换。作为概念证明,我们在八个 RBS、八个启动子和四个复制起点的控制下构建了多个sfGFP表达载体,以及表达黑色素原紫草素生物合成基因的诱导型四基因操纵子。为了证明 SEVA 3.1 载体的互操作性,所有构建体都在恶臭假单胞菌和大肠杆菌中进行了表征。总之,SEVA 3.1 平台通过成本和时间友好的 DNA 组装方法优化了插入物和主链的兼容性和模块化,大大扩展了革兰氏阴性细菌中成功合成生物学应用的工具箱。











































 京公网安备 11010802027423号
京公网安备 11010802027423号