当前位置:
X-MOL 学术
›
Bull. Korean Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Balancing All‐Atom Force Field for DNA Simulations Using Osmotic Pressure Data
Bulletin of the Korean Chemical Society ( IF 2.3 ) Pub Date : 2020-07-06 , DOI: 10.1002/bkcs.12065 Hyeonjun Kim 1 , Youngshang Pak 1
Bulletin of the Korean Chemical Society ( IF 2.3 ) Pub Date : 2020-07-06 , DOI: 10.1002/bkcs.12065 Hyeonjun Kim 1 , Youngshang Pak 1
Affiliation
|
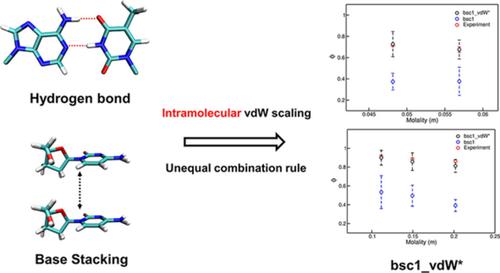
For classical molecular dynamics (MD) simulations of DNA molecules, several all‐atom force fields have been developed, including the very popular amber bsc1 force field. Despite marked improvement from the standard amber99 force field, this force field has a tendency to underestimate base‐pair hydrogen bonds and overestimate base stacking (BS). Previously, the weakened hydrogen bond with the bsc1 was improved by modulating a limited number of van der Waals sigma parameters. In this study, to resolve the remaining issue of overestimating BS, nonbonding interaction parameters relevant to the BS interaction were tuned, such that the experimental osmotic pressure coefficients of deoxy‐ribonucleoside solutions are closely reproduced. This newly modified force field is more balanced in terms of nonbonding interactions and can be applied to many challenging computational problems of DNA folding and breathing.
中文翻译:

使用渗透压数据平衡DNA模拟的全原子力场
对于DNA分子的经典分子动力学(MD)模拟,已经开发了几种全原子力场,包括非常流行的琥珀色bsc1力场。尽管从标准的amber99力场获得了显着改善,但该力场仍倾向于低估碱基对氢键,而高估了碱基堆积(BS)。以前,通过调节有限数量的范德华σ参数可以改善bsc1弱的氢键。在这项研究中,为解决剩余的高估BS的问题,调整了与BS相互作用相关的非键相互作用参数,从而精确再现了脱氧核糖核苷溶液的实验渗透压系数。
更新日期:2020-07-06
中文翻译:

使用渗透压数据平衡DNA模拟的全原子力场
对于DNA分子的经典分子动力学(MD)模拟,已经开发了几种全原子力场,包括非常流行的琥珀色bsc1力场。尽管从标准的amber99力场获得了显着改善,但该力场仍倾向于低估碱基对氢键,而高估了碱基堆积(BS)。以前,通过调节有限数量的范德华σ参数可以改善bsc1弱的氢键。在这项研究中,为解决剩余的高估BS的问题,调整了与BS相互作用相关的非键相互作用参数,从而精确再现了脱氧核糖核苷溶液的实验渗透压系数。











































 京公网安备 11010802027423号
京公网安备 11010802027423号