Cell Systems ( IF 9.0 ) Pub Date : 2020-07-24 , DOI: 10.1016/j.cels.2020.06.013 Mi Yang 1 , Francesca Petralia 2 , Zhi Li 3 , Hongyang Li 4 , Weiping Ma 2 , Xiaoyu Song 5 , Sunkyu Kim 6 , Heewon Lee 6 , Han Yu 7 , Bora Lee 8 , Seohui Bae 9 , Eunji Heo 10 , Jan Kaczmarczyk 11 , Piotr Stępniak 11 , Michał Warchoł 11 , Thomas Yu 12 , Anna P Calinawan 2 , Paul C Boutros 13 , Samuel H Payne 14 , Boris Reva 2 , , Emily Boja 15 , Henry Rodriguez 15 , Gustavo Stolovitzky 16 , Yuanfang Guan 4 , Jaewoo Kang 6 , Pei Wang 2 , David Fenyö 3 , Julio Saez-Rodriguez 17
|
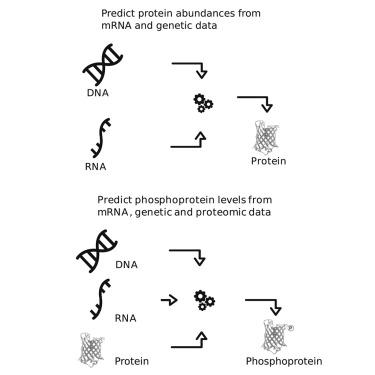
Cancer is driven by genomic alterations, but the processes causing this disease are largely performed by proteins. However, proteins are harder and more expensive to measure than genes and transcripts. To catalyze developments of methods to infer protein levels from other omics measurements, we leveraged crowdsourcing via the NCI-CPTAC DREAM proteogenomic challenge. We asked for methods to predict protein and phosphorylation levels from genomic and transcriptomic data in cancer patients. The best performance was achieved by an ensemble of models, including as predictors transcript level of the corresponding genes, interaction between genes, conservation across tumor types, and phosphosite proximity for phosphorylation prediction. Proteins from metabolic pathways and complexes were the best and worst predicted, respectively. The performance of even the best-performing model was modest, suggesting that many proteins are strongly regulated through translational control and degradation. Our results set a reference for the limitations of computational inference in proteogenomics.
A record of this paper’s transparent peer review process is included in the Supplemental Information.
中文翻译:

从基因组学和转录组学对癌症蛋白质和磷蛋白水平的可预测性进行社区评估。
癌症是由基因组改变驱动的,但导致这种疾病的过程主要由蛋白质执行。然而,蛋白质比基因和转录物更难测量,也更昂贵。为了促进从其他组学测量中推断蛋白质水平的方法的发展,我们通过 NCI-CPTAC DREAM 蛋白质组学挑战利用众包。我们要求从癌症患者的基因组和转录组数据中预测蛋白质和磷酸化水平的方法。最好的性能是由一组模型实现的,包括作为预测因子的相应基因的转录水平、基因之间的相互作用、跨肿瘤类型的保守性以及用于磷酸化预测的磷酸化位点接近度。来自代谢途径和复合物的蛋白质分别是最好的和最差的预测。即使是性能最好的模型的性能也不高,这表明许多蛋白质通过翻译控制和降解受到强烈调节。我们的结果为蛋白质基因组学中计算推理的局限性提供了参考。
本文的透明同行评审过程的记录包含在补充信息中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号