当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Splicing impact of deep exonic missense variants in CAPN3 explored systematically by minigene functional assay.
Human Mutation ( IF 3.3 ) Pub Date : 2020-07-15 , DOI: 10.1002/humu.24083 Eugénie Dionnet 1 , Aurélia Defour 1 , Nathalie Da Silva 1 , Alexandra Salvi 1 , Nicolas Lévy 1, 2, 3 , Martin Krahn 1, 2 , Marc Bartoli 1 , Francesca Puppo 1 , Svetlana Gorokhova 1, 2
Human Mutation ( IF 3.3 ) Pub Date : 2020-07-15 , DOI: 10.1002/humu.24083 Eugénie Dionnet 1 , Aurélia Defour 1 , Nathalie Da Silva 1 , Alexandra Salvi 1 , Nicolas Lévy 1, 2, 3 , Martin Krahn 1, 2 , Marc Bartoli 1 , Francesca Puppo 1 , Svetlana Gorokhova 1, 2
Affiliation
|
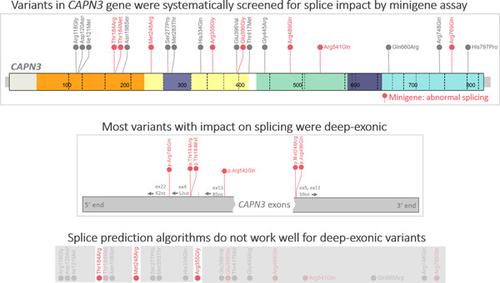
Improving the accuracy of variant interpretation during diagnostic sequencing is a major goal for genomic medicine. To explore an often‐overlooked splicing effect of missense variants, we developed the functional assay (“minigene”) for the majority of exons of CAPN3, the gene responsible for limb girdle muscular dystrophy. By systematically screening 21 missense variants distributed along the gene, we found that eight clinically relevant missense variants located at a certain distance from the exon–intron borders (deep exonic missense variants) disrupted normal splicing of CAPN3 exons. Several recent machine learning‐based computational tools failed to predict splicing impact for the majority of these deep exonic missense variants, highlighting the importance of including variants of this type in the training sets during the future algorithm development. Overall, 24 variants in CAPN3 gene were explored, leading to the change in the American College of Medical Genetics and Genomics classification of seven of them when results of the “minigene” functional assay were considered. Our findings reveal previously unknown splicing impact of several clinically important variants in CAPN3 and draw attention to the existence of deep exonic variants with a disruptive effect on gene splicing that could be overlooked by the current approaches in clinical genetics.
中文翻译:

通过小基因功能测定系统探索了 CAPN3 中深外显子错义变体的剪接影响。
在诊断测序期间提高变异解释的准确性是基因组医学的主要目标。为了探索错义变体经常被忽视的剪接效应,我们开发了CAPN3 大部分外显子的功能测定(“小基因”),CAPN3是导致肢带型肌营养不良症的基因。通过系统筛选沿基因分布的 21 个错义变体,我们发现位于外显子-内含子边界一定距离处的 8 个临床相关错义变体(深外显子错义变体)破坏了CAPN3的正常剪接外显子。最近一些基于机器学习的计算工具未能预测大多数这些深度外显子错义变体的剪接影响,突出了在未来算法开发过程中将这种类型的变体包含在训练集中的重要性。总体而言,研究了CAPN3基因的 24 个变体,当考虑到“小基因”功能测定的结果时,导致美国医学遗传学和基因组学学院对其中 7 个的分类发生了变化。我们的研究结果揭示了CAPN3中几个临床上重要的变体以前未知的剪接影响,并引起人们对深外显子变体的存在的关注,这些变体对基因剪接具有破坏性影响,这可能被当前临床遗传学方法所忽视。
更新日期:2020-07-15
中文翻译:

通过小基因功能测定系统探索了 CAPN3 中深外显子错义变体的剪接影响。
在诊断测序期间提高变异解释的准确性是基因组医学的主要目标。为了探索错义变体经常被忽视的剪接效应,我们开发了CAPN3 大部分外显子的功能测定(“小基因”),CAPN3是导致肢带型肌营养不良症的基因。通过系统筛选沿基因分布的 21 个错义变体,我们发现位于外显子-内含子边界一定距离处的 8 个临床相关错义变体(深外显子错义变体)破坏了CAPN3的正常剪接外显子。最近一些基于机器学习的计算工具未能预测大多数这些深度外显子错义变体的剪接影响,突出了在未来算法开发过程中将这种类型的变体包含在训练集中的重要性。总体而言,研究了CAPN3基因的 24 个变体,当考虑到“小基因”功能测定的结果时,导致美国医学遗传学和基因组学学院对其中 7 个的分类发生了变化。我们的研究结果揭示了CAPN3中几个临床上重要的变体以前未知的剪接影响,并引起人们对深外显子变体的存在的关注,这些变体对基因剪接具有破坏性影响,这可能被当前临床遗传学方法所忽视。











































 京公网安备 11010802027423号
京公网安备 11010802027423号