Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2020-07-15 , DOI: 10.1016/j.csbj.2020.07.004 Zeju Sun 1 , Shaojun Pei 1 , Rong Lucy He 2 , Stephen S-T Yau 1
|
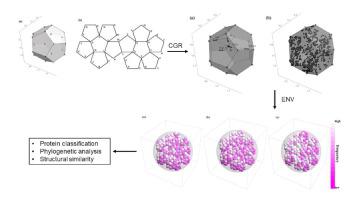
Chaos Game Representation (CGR) was first proposed to be an image representation method of DNA and have been extended to the case of other biological macromolecules. Compared with the CGR images of DNA, where DNA sequences are converted into a series of points in the unit square, the existing CGR images of protein are not so elegant in geometry and the implications of the distribution of points in the CGR image are not so obvious. In this study, by naturally distributing the twenty amino acids on the vertices of a regular dodecahedron, we introduce a novel three-dimensional image representation of protein sequences with CGR method. We also associate each CGR image with a vector in high dimensional Euclidean space, called the extended natural vector (ENV), in order to analyze the information contained in the CGR images. Based on the results of protein classification and phylogenetic analysis, our method could serve as a precise method to discover biological relationships between proteins.
中文翻译:

蛋白质的新型数值表示法:三维混沌博弈表示法及其扩展的自然向量。
混沌博弈表示法(CGR)最初被提出是DNA的图像表示方法,并已扩展到其他生物大分子的情况。与DNA的CGR图像相比,DNA序列被转换为单位正方形中的一系列点,而现有的蛋白质CGR图像的几何形状并不那么优雅,并且CGR图像中点分布的含义也不那么优雅明显。在这项研究中,通过在规则十二面体的顶点上自然分布二十个氨基酸,我们用CGR方法介绍了蛋白质序列的新型三维图像表示。我们还将每个CGR图像与高维欧氏空间中的矢量(称为扩展自然矢量(ENV))相关联,以分析CGR图像中包含的信息。



























 京公网安备 11010802027423号
京公网安备 11010802027423号