Cell Systems ( IF 9.0 ) Pub Date : 2020-07-14 , DOI: 10.1016/j.cels.2020.06.005 Shilpa Nadimpalli Kobren 1 , Bernard Chazelle 2 , Mona Singh 3
|
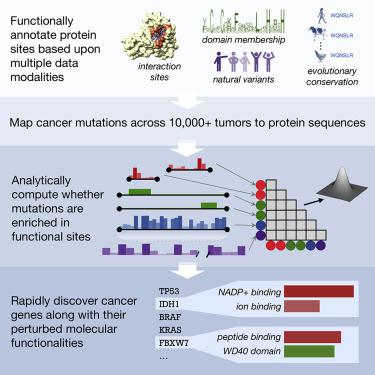
A major challenge in cancer genomics is to identify genes with functional roles in cancer and uncover their mechanisms of action. We introduce an integrative framework that identifies cancer-relevant genes by pinpointing those whose interaction or other functional sites are enriched in somatic mutations across tumors. We derive analytical calculations that enable us to avoid time-prohibitive permutation-based significance tests, making it computationally feasible to simultaneously consider multiple measures of protein site functionality. Our accompanying software, PertInInt, combines knowledge about sites participating in interactions with DNA, RNA, peptides, ions, or small molecules with domain, evolutionary conservation, and gene-level mutation data. When applied to 10,037 tumor samples, PertInInt uncovers both known and newly predicted cancer genes, while additionally revealing what types of interactions or other functionalities are disrupted. PertInInt's analysis demonstrates that somatic mutations are frequently enriched in interaction sites and domains and implicates interaction perturbation as a pervasive cancer-driving event.
中文翻译:

PertInInt:一种快速发现具有扰动相互作用和功能的癌症驱动基因的综合分析方法。
癌症基因组学的一个主要挑战是识别在癌症中具有功能作用的基因并揭示它们的作用机制。我们引入了一个综合框架,通过确定那些相互作用或其他功能位点在整个肿瘤中富含体细胞突变的基因来识别癌症相关基因。我们推导出分析计算,使我们能够避免基于时间禁止排列的显着性测试,从而使同时考虑蛋白质位点功能的多种测量在计算上可行。我们随附的软件 PertInInt 将有关参与 DNA、RNA、肽、离子或小分子相互作用的位点的知识与域、进化保守性和基因级突变数据相结合。当应用于 10,037 个肿瘤样本时,PertInInt 揭示了已知和新预测的癌症基因,同时还揭示了哪些类型的相互作用或其他功能被破坏。PertInInt 的分析表明,体细胞突变经常在相互作用位点和域中丰富,并将相互作用扰动作为普遍存在的癌症驱动事件。











































 京公网安备 11010802027423号
京公网安备 11010802027423号