Journal of Asia-Pacific Entomology ( IF 1.1 ) Pub Date : 2020-07-13 , DOI: 10.1016/j.aspen.2020.07.001 Avas Pakrashi , Shantanu Kundu , Jagdish Saini , Kaomud Tyagi , Kailash Chandra , Vikas Kumar
|
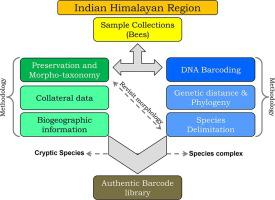
DNA barcoding has largely been tested for a wide range of taxa and evidenced as a reliable and rapid molecular tool for species-level identification. The present study lends to generate 156 DNA barcodes, of which 141 belonged to 30 morphologically identified bees from the Indian Himalayan Regions (IHRs). The generated barcode data along with 84 sequences of global database distinctly discriminated all the studied species with sufficient genetic distances and cohesive monophyletic clustering in Bayesian analysis (BA) phylogeny. The species delimitation methods, Automatic Barcode Gap Discovery (ABGD), Bayesian Poisson-Tree-Processes (bPTP), and General Mixed Yule-coalescent (GMYC) yielded 68, 70, and 71 molecular operational taxonomic units (MOTUs) respectively. The present DNA barcode-based examination detected the possible cryptic diversity in two Apis species (A. cerana and A. dorsata), Bombus hypnorum, Lepidotrigona arcifera, and Ceratina sutepensis. The present study also evidenced the species complexes within Bombus albopleuralis and Bombus trifasciatus in the IHRs. The species delimitation methods also detected an additional seven putative species from the IHRs, which were identified up to the genus level. In conclusion, this preliminary effort helps to develop a reliable barcode database of bees from the Indian IHRs to facilitate the future systematics study. These molecular data can be utilized to evaluate the population structures and assist to formulate the effective plans for bee conservation.
中文翻译:

从印度喜马拉雅山中选择蜜蜂的分子鉴定:初步工作
DNA条形码已针对各种分类单元进行了广泛的测试,并被证明是用于物种级别识别的可靠且快速的分子工具。本研究有助于产生156个DNA条形码,其中141个属于来自印度喜马拉雅地区(IHR)的30株形态鉴定的蜜蜂。所生成的条形码数据以及全球数据库的84个序列,在贝叶斯分析(BA)系统发育中,以足够的遗传距离和具有凝聚力的单系聚类,可以明显地区分所有被研究物种。物种定界方法,自动条形码差距发现(ABGD),贝叶斯泊松树过程(bPTP)和通用混合尤尔柱盟(GMYC)分别产生了68、70和71个分子操作分类单位(MOTU)。A. cerana和A. dorsata),Bombus hypnorum,Lepidotrigona arcifera和Ceratina sutepensis。本研究还证明了《国际卫生条例》中的白腹蓬和黑腹蓬中的物种复合体。物种划界方法还从《国际卫生条例》中检测出了另外七个推定物种,这些物种被鉴定到属水平。总之,这项初步工作有助于从印度《国际卫生条例》建立可靠的蜜蜂条形码数据库,以促进未来的系统研究。这些分子数据可用于评估种群结构,并协助制定有效的养蜂计划。











































 京公网安备 11010802027423号
京公网安备 11010802027423号