Journal of Structural Biology ( IF 3.0 ) Pub Date : 2020-07-10 , DOI: 10.1016/j.jsb.2020.107559 Azat Gabdulkhakov 1 , Ivan Mitroshin 1 , Maria Garber 1
|
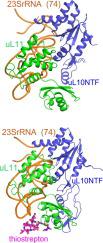
Complexes of archaeal ribosomal proteins uL11 and uL10/P0 (the two-domain N-terminal fragment of uL10, uL10NTF/P0NTF) with the adjacent 74 nucleotides of 23S rRNA fragment (23SrRNA(74)) from Methanococcus jannaschii (Mja) were obtained, crystallized and their structures were studied. The comparative structural analysis of the complexes of Mja uL10NTF•23SrRNA(74) and Mja uL10NTF•uL11•23SrRNA(74) shows that the insertion of uL11 in the binary complex does not change the conformation of the 23S rRNA fragment. On the other hand, the interaction with this specific RNA fragment leads to the restructuring of uL11 compared to the structure of this protein in the free state. Besides, although analysis confirmed the mobility of uL10/P0 domain II, disproved the assumption that it may be in contact with rRNA or uL11. In addition, the Mja uL10NTF•uL11•23SrRNA(74) complex was cocrystallized with the antibiotic thiostrepton, and the structure of this complex was solved. The thiostrepton binding site in this archaeal complex was found between the 23S rRNA and the N-terminal domain (NTD) of the Mja uL11 protein, similar to its binding site in the one of bacterial ribosome complex with thiostrepton. Upon binding of thiostrepton, the NTD of uL11 shifts toward rRNA by 7 Å. Such a shift may be the cause of the inhibitory effect of the antibiotic on the recruitment of translation factors to the GTPase-activating region in archaeal ribosomes, similar to its inhibitory effect on protein synthesis in bacterial ribosomes.
中文翻译:

古细菌詹氏甲烷球菌中核糖体 P 茎基的结构。
古细菌核糖体蛋白 uL11 和 uL10/P0(uL10 的两个结构域 N 端片段,uL10NTF/P0NTF)与来自詹氏甲烷球菌的 23S rRNA 片段(23SrRNA(74))的相邻 74 个核苷酸的复合物(Mja) 被获得,结晶并研究了它们的结构。Mja uL10NTF•23SrRNA(74)和Mja uL10NTF•uL11•23SrRNA(74)复合物的结构比较分析表明,在二元复合物中插入uL11并没有改变23S rRNA片段的构象。另一方面,与此蛋白质在游离状态下的结构相比,与此特定 RNA 片段的相互作用导致 uL11 的重组。此外,虽然分析证实了 uL10/P0 域 II 的移动性,但反驳了它可能与 rRNA 或 uL11 接触的假设。此外,Mja uL10NTF•uL11•23SrRNA(74)复合物与抗生素硫链丝菌素共结晶,并解析了该复合物的结构。该古菌复合物中的硫链丝菌肽结合位点位于 23S rRNA 和 Mja uL11 蛋白的 N 端结构域 (NTD) 之间,类似于其在细菌核糖体与硫链丝菌素复合物中的结合位点。与硫链丝菌素结合后,uL11 的 NTD 向 rRNA 移动 7 Å。这种转变可能是抗生素对古细菌核糖体中 GTPase 激活区翻译因子募集的抑制作用的原因,类似于其对细菌核糖体中蛋白质合成的抑制作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号