Journal of Structural Biology ( IF 3.0 ) Pub Date : 2020-07-10 , DOI: 10.1016/j.jsb.2020.107575 Jeddidiah W D Griffin 1
|
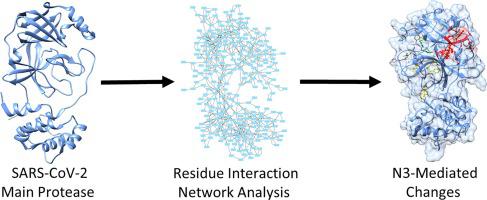
COVID-19 is a respiratory disease caused by the coronavirus SARS-CoV-2. SARS-CoV-2 has many similarities with SARS-CoV. Both viruses rely on a protease called the main protease, or Mpro, for replication. Therefore, inhibiting Mpro may be a successful strategy for treating COVID-19. Structures of the main proteases of SARS-CoV and SARS-CoV-2 with and without inhibitor N3 are available in the Protein Data Bank. Comparing these structures revealed residue interaction network changes associated with N3 inhibition. Comparing network clustering with and without inhibitor N3 identified the formation of a cluster of residues 17, 18, 30–33, 70, 95, 98, 103, 117, 122, and 177 as a network change in both viral proteases when bound to inhibitor N3. Betweenness and stress centrality differences as well as differences in bond energies and relative B-factors when comparing free Mpro to inhibitor-bound Mpro identified residues 131, 175, 182, and 185 as possibly conformationally relevant when bound to the inhibitor N3. Taken together, these results provide insight into conformational changes of betacoronavirus Mpros when bound to an inhibitor.
中文翻译:

当结合抑制剂N3时,SARS-CoV和SARS-CoV-2主要蛋白酶残基相互作用网络发生变化。
COVID-19是由冠状病毒SARS-CoV-2引起的呼吸道疾病。SARS-CoV-2与SARS-CoV有很多相似之处。两种病毒都依赖一种称为主蛋白酶或M pro的蛋白酶进行复制。因此,抑制M pro可能是治疗COVID-19的成功策略。蛋白质数据库中有和没有抑制剂N3的SARS-CoV和SARS-CoV-2主要蛋白酶的结构。比较这些结构揭示了与N3抑制相关的残基相互作用网络的变化。比较有抑制剂和无抑制剂N3的网络聚类,发现与抑制剂结合时,两种病毒蛋白酶的网络变化都形成了残基17、18、30-33、70、95、98、103、117、122和177的簇N3 比较游离M pro与抑制剂结合的M pro时,中间度和应力中心性差异以及键能和相对B因子的差异当与抑制剂N3结合时,鉴定出的残基131、175、182和185可能在构象上相关。综上所述,这些结果提供了与抑制剂结合时β冠状病毒M pro构象变化的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号