当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Increasing the Coverage of a Mass Spectral Library of Milk Oligosaccharides Using a Hybrid-Search-Based Bootstrapping Method and Milks from a Wide Variety of Mammals.
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-07-08 , DOI: 10.1021/acs.analchem.0c00342 Concepcion Africano Remoroza 1 , Yuxue Liang 1 , Tytus D Mak 1 , Yuri Mirokhin 1 , Sergey L Sheetlin 1 , Xiaoyu Yang 1 , Joice V San Andres 2, 3 , Michael L Power 4 , Stephen E Stein 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-07-08 , DOI: 10.1021/acs.analchem.0c00342 Concepcion Africano Remoroza 1 , Yuxue Liang 1 , Tytus D Mak 1 , Yuri Mirokhin 1 , Sergey L Sheetlin 1 , Xiaoyu Yang 1 , Joice V San Andres 2, 3 , Michael L Power 4 , Stephen E Stein 1
Affiliation
|
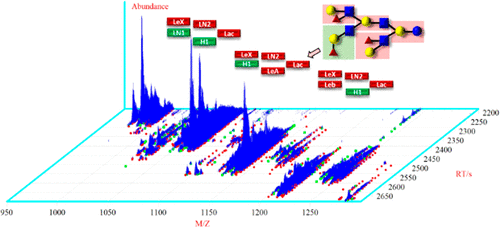
This study significantly expands both the scope and method of identification for construction of a previously reported tandem mass spectral library of 74 human milk oligosaccharides (HMOs) derived from results of combined LC-MS/MS experiments and comprehensive structural analysis of HMOs. In the present work, a hybrid search “bootstrap” identification method was employed that substantially broadens the coverage of milk oligosaccharides and thereby increases utility use of a spectrum library-based method for the rapid tentative identification of all distinguishable glycans in milk. This involved hybrid searching of the previous library, which was itself constructed using the hybrid search of oligosaccharide spectra in the NIST 17 Tandem MS Library. The general approach appears applicable to library construction of other classes of compounds. The coverage of oligosaccharides was significantly extended using milks from a variety of mammals, including bovine, Asian buffalo, African lion, and goat. This new method led to the identification of another 145 oligosaccharides, including an additional 80 HMOs from reanalysis of human milk. The newly identified compounds were added to a freely available mass spectral reference database of 219 milk oligosaccharides. We also provide suggestions to overcome several limitations and pitfalls in the interpretation of spectra of unknown oligosaccharides.
中文翻译:

使用基于混合搜索的自举方法和来自多种哺乳动物的牛奶,增加牛奶寡糖质谱库的覆盖率。
这项研究显着扩展了构建先前报道的74种人乳寡糖(HMO)串联质谱库的鉴定范围和方法,这些库是从LC-MS / MS组合实验和HMO的综合结构分析结果得出的。在本工作中,采用了一种混合搜索“ bootstrap”识别方法,该方法大大拓宽了牛奶寡糖的覆盖范围,从而提高了基于光谱库的方法对牛奶中所有可区分聚糖的快速初步鉴定的实用性。这涉及对先前库的混合搜索,该库本身是使用NIST 17串联MS库中的寡糖谱图进行混合搜索构建的。通用方法似乎适用于其他类别化合物的文库构建。使用包括牛,亚洲水牛,非洲狮和山羊在内的多种哺乳动物的乳汁,寡糖的覆盖范围得到了显着扩展。这种新方法导致了对另外145种寡糖的鉴定,其中包括通过对人乳的重新分析获得的另外80种HMO。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。
更新日期:2020-08-04
中文翻译:

使用基于混合搜索的自举方法和来自多种哺乳动物的牛奶,增加牛奶寡糖质谱库的覆盖率。
这项研究显着扩展了构建先前报道的74种人乳寡糖(HMO)串联质谱库的鉴定范围和方法,这些库是从LC-MS / MS组合实验和HMO的综合结构分析结果得出的。在本工作中,采用了一种混合搜索“ bootstrap”识别方法,该方法大大拓宽了牛奶寡糖的覆盖范围,从而提高了基于光谱库的方法对牛奶中所有可区分聚糖的快速初步鉴定的实用性。这涉及对先前库的混合搜索,该库本身是使用NIST 17串联MS库中的寡糖谱图进行混合搜索构建的。通用方法似乎适用于其他类别化合物的文库构建。使用包括牛,亚洲水牛,非洲狮和山羊在内的多种哺乳动物的乳汁,寡糖的覆盖范围得到了显着扩展。这种新方法导致了对另外145种寡糖的鉴定,其中包括通过对人乳的重新分析获得的另外80种HMO。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。新鉴定的化合物被添加到219种牛奶寡糖的免费质谱参考数据库中。我们还提供了一些建议,以克服未知寡糖光谱解释中的一些限制和陷阱。



























 京公网安备 11010802027423号
京公网安备 11010802027423号