当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Benchmarking 2D/3D/MD-QSAR Models for Imatinib Derivatives: How Far Can We Predict?
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-07-05 , DOI: 10.1021/acs.jcim.0c00200 Phyo Phyo Kyaw Zin 1 , Alexandre Borrel 1 , Denis Fourches 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-07-05 , DOI: 10.1021/acs.jcim.0c00200 Phyo Phyo Kyaw Zin 1 , Alexandre Borrel 1 , Denis Fourches 1
Affiliation
|
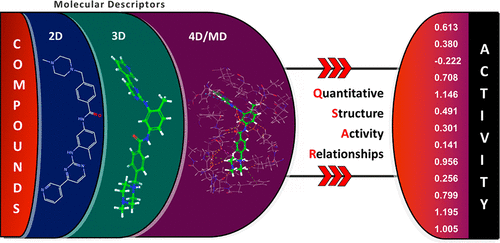
Imatinib, a 2-phenylaminopyridine-based BCR-ABL tyrosine kinase inhibitor, is a highly effective drug for treating Chronic Myeloid Leukemia (CML). However, cases of drug resistance are constantly emerging due to various mutations in the ABL kinase domain; thus, it is crucial to identify novel bioactive analogues. Reliable QSAR models and molecular docking protocols have been shown to facilitate the discovery of new compounds from chemical libraries prior to experimental testing. However, as the vast majority of QSAR models strictly relies on 2D descriptors, the rise of 3D descriptors directly computed from molecular dynamics simulations offers new opportunities to potentially augment the reliability of QSAR models. Herein, we employed molecular docking and molecular dynamics on a large series of Imatinib derivatives and developed an ensemble of QSAR models relying on deep neural nets (DNN) and hybrid sets of 2D/3D/MD descriptors in order to predict the binding affinity and inhibition potencies of those compounds. Through rigorous validation tests, we showed that our DNN regression models achieved excellent external prediction performances for the pKi data set (n = 555, R2 ≥ 0.71. and MAE ≤ 0.85), and the pIC50 data set (n = 306, R2 ≥ 0.54. and MAE ≤ 0.71) with strict validation protocols based on external test sets and 10-fold native and nested cross validations. Interestingly, the best DNN and random forest models performed similarly across all descriptor sets. In fact, for this particular series of compounds, our external test results suggest that incorporating additional 3D protein–ligand binding site fingerprint, descriptors, or even MD time-series descriptors did not significantly improve the overall R2 but lowered the MAE of DNN QSAR models. Those augmented models could still help in identifying and understanding the key dynamic protein–ligand interactions to be optimized for further molecular design.
中文翻译:

对伊马替尼衍生物的2D / 3D / MD-QSAR模型进行基准测试:我们可以预测多远?
伊马替尼是一种基于2-苯基氨基吡啶的BCR-ABL酪氨酸激酶抑制剂,是治疗慢性粒细胞白血病(CML)的高效药物。然而,由于ABL激酶域中的各种突变,耐药性病例不断出现。因此,鉴定新型生物活性类似物至关重要。可靠的QSAR模型和分子对接方案已被证明有助于在实验测试之前从化学文库中发现新化合物。但是,由于绝大多数QSAR模型严格依赖于2D描述子,因此直接从分子动力学模拟计算得出的3D描述子的兴起为潜在地提高QSAR模型的可靠性提供了新的机会。在这里 我们在大量的伊马替尼衍生物上应用了分子对接和分子动力学,并开发了依赖于深层神经网络(DNN)和2D / 3D / MD描述子混合集的QSAR模型集合,以预测其结合亲和力和抑制力这些化合物。通过严格的验证测试,我们证明了DNN回归模型对pKi数据集具有出色的外部预测性能(n = 555,R2 ≥0.71。和MAE≤0.85),和PIC 50数据集(N = 306,R 2 ≥0.54。和MAE≤0.71)与基于外部测试集和10倍天然和嵌套交叉验证严格验证协议。有趣的是,最佳DNN和随机森林模型在所有描述符集上的表现相似。实际上,对于该特定系列的化合物,我们的外部测试结果表明,结合其他3D蛋白-配体结合位点指纹,描述符甚至MD时间序列描述符并不能显着改善总体R 2,但降低了DNN QSAR的MAE楷模。这些增强的模型仍然可以帮助识别和理解关键的动态蛋白质-配体相互作用,以进一步优化分子设计。
更新日期:2020-07-27
中文翻译:

对伊马替尼衍生物的2D / 3D / MD-QSAR模型进行基准测试:我们可以预测多远?
伊马替尼是一种基于2-苯基氨基吡啶的BCR-ABL酪氨酸激酶抑制剂,是治疗慢性粒细胞白血病(CML)的高效药物。然而,由于ABL激酶域中的各种突变,耐药性病例不断出现。因此,鉴定新型生物活性类似物至关重要。可靠的QSAR模型和分子对接方案已被证明有助于在实验测试之前从化学文库中发现新化合物。但是,由于绝大多数QSAR模型严格依赖于2D描述子,因此直接从分子动力学模拟计算得出的3D描述子的兴起为潜在地提高QSAR模型的可靠性提供了新的机会。在这里 我们在大量的伊马替尼衍生物上应用了分子对接和分子动力学,并开发了依赖于深层神经网络(DNN)和2D / 3D / MD描述子混合集的QSAR模型集合,以预测其结合亲和力和抑制力这些化合物。通过严格的验证测试,我们证明了DNN回归模型对pKi数据集具有出色的外部预测性能(n = 555,R2 ≥0.71。和MAE≤0.85),和PIC 50数据集(N = 306,R 2 ≥0.54。和MAE≤0.71)与基于外部测试集和10倍天然和嵌套交叉验证严格验证协议。有趣的是,最佳DNN和随机森林模型在所有描述符集上的表现相似。实际上,对于该特定系列的化合物,我们的外部测试结果表明,结合其他3D蛋白-配体结合位点指纹,描述符甚至MD时间序列描述符并不能显着改善总体R 2,但降低了DNN QSAR的MAE楷模。这些增强的模型仍然可以帮助识别和理解关键的动态蛋白质-配体相互作用,以进一步优化分子设计。











































 京公网安备 11010802027423号
京公网安备 11010802027423号