当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
mycoCSM: Using Graph-Based Signatures to Identify Safe Potent Hits against Mycobacteria.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-07-02 , DOI: 10.1021/acs.jcim.0c00362 Douglas E V Pires 1, 2, 3 , David B Ascher 1, 2, 4
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-07-02 , DOI: 10.1021/acs.jcim.0c00362 Douglas E V Pires 1, 2, 3 , David B Ascher 1, 2, 4
Affiliation
|
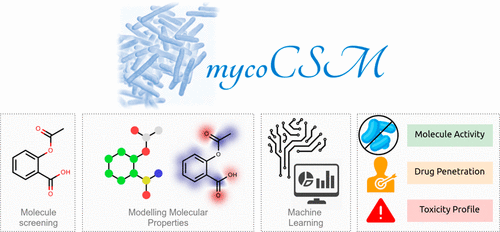
Development of new potent, safe drugs to treat Mycobacteria has proven to be challenging, with limited hit rates of initial screens restricting subsequent development efforts. Despite significant efforts and the evolution of quantitative structure–activity relationship as well as machine learning-based models for computationally predicting molecule bioactivity, there is an unmet need for efficient and reliable methods for identifying biologically active compounds against Mycobacterium that are also safe for humans. Here we developed mycoCSM, a graph-based signature approach to rapidly identify compounds likely to be active against bacteria from the genus Mycobacterium, or against specific Mycobacteria species. mycoCSM was trained and validated on eight organism-specific and for the first time a general Mycobacteria data set, achieving correlation coefficients of up to 0.89 on cross-validation and 0.88 on independent blind tests, when predicting bioactivity in terms of minimum inhibitory concentration. In addition, we also developed a predictor to identify those compounds likely to penetrate in necrotic tuberculosis foci, which achieved a correlation coefficient of 0.75. Together with a built-in estimator of the maximum tolerated dose in humans, we believe this method will provide a valuable resource to enrich screening libraries with potent, safe molecules. To provide simple guidance in the selection of libraries with favorable anti-Mycobacteria properties, we made mycoCSM freely available online at http://biosig.unimelb.edu.au/myco_csm.
中文翻译:

mycoCSM:使用基于图的签名来确定针对分枝杆菌的安全有效命中。
事实证明,开发治疗分枝杆菌的新型有效,安全的药物具有挑战性,初始筛选的成功率有限,从而限制了后续的开发工作。尽管付出了巨大的努力,并且定量构效关系的发展以及基于机器学习的模型可预测分子的生物活性,但仍需要有效而可靠的方法来鉴定对分枝杆菌也对人类安全的生物活性化合物。在这里,我们开发了mycoCSM,这是一种基于图形的签名方法,可以快速识别可能对分枝杆菌属细菌或特定分枝杆菌物种具有活性的化合物。对mycoCSM进行了八种特定于生物体的培训和验证,这是首次针对一般分枝杆菌数据集,当根据最小抑菌浓度预测生物活性时,交叉验证的相关系数高达0.89,独立盲测的相关系数高达0.88。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。当以最小抑菌浓度预测生物活性时。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。当以最小抑菌浓度预测生物活性时。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。
更新日期:2020-07-27
中文翻译:

mycoCSM:使用基于图的签名来确定针对分枝杆菌的安全有效命中。
事实证明,开发治疗分枝杆菌的新型有效,安全的药物具有挑战性,初始筛选的成功率有限,从而限制了后续的开发工作。尽管付出了巨大的努力,并且定量构效关系的发展以及基于机器学习的模型可预测分子的生物活性,但仍需要有效而可靠的方法来鉴定对分枝杆菌也对人类安全的生物活性化合物。在这里,我们开发了mycoCSM,这是一种基于图形的签名方法,可以快速识别可能对分枝杆菌属细菌或特定分枝杆菌物种具有活性的化合物。对mycoCSM进行了八种特定于生物体的培训和验证,这是首次针对一般分枝杆菌数据集,当根据最小抑菌浓度预测生物活性时,交叉验证的相关系数高达0.89,独立盲测的相关系数高达0.88。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。当以最小抑菌浓度预测生物活性时。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。当以最小抑菌浓度预测生物活性时。此外,我们还开发了一种预测因子,以鉴定可能穿透坏死结核病灶的那些化合物,相关系数达到0.75。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。结合内置的人类最大耐受剂量估算器,我们相信该方法将为宝贵的资源提供有用的,安全的分子来丰富筛选库。为了在选择具有良好抗分枝杆菌特性的文库时提供简单指导,我们在http://biosig.unimelb.edu.au/myco_csm上免费在线提供了mycoCSM。











































 京公网安备 11010802027423号
京公网安备 11010802027423号