当前位置:
X-MOL 学术
›
Anal. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A QCM-based rupture event scanning technique as a simple and reliable approach to study the kinetics of DNA duplex dissociation.
Analytical Methods ( IF 2.7 ) Pub Date : 2020-07-02 , DOI: 10.1039/d0ay00613k N N Kurus 1 , F N Dultsev , V M Golyshev , D V Nekrasov , D V Pyshnyi , A A Lomzov
Analytical Methods ( IF 2.7 ) Pub Date : 2020-07-02 , DOI: 10.1039/d0ay00613k N N Kurus 1 , F N Dultsev , V M Golyshev , D V Nekrasov , D V Pyshnyi , A A Lomzov
Affiliation
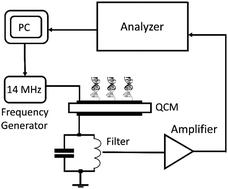
|
Rupture Event Scanning (REVS) is applied for the first time within an approach based on dynamic force spectroscopy. Using model DNA duplexes containing 20 pairs of oligonucleotides including those containing single mismatches, we demonstrated the possibility of reliable determination of the kinetic parameters of dissociation of biomolecular complexes: barrier positions, the rate constants of dissociation, and the lifetime of complexes. Within this approach, mechanical dissociation of DNA duplexes occurs according to a mechanism similar to unzipping. It is shown that this process takes place by overcoming a single energy barrier. In the case where a mismatch is located at the farthest duplex end from the QCM surface, a substantial decrease in the position of the barrier between the bound and unbound states is observed. We suppose that this is due to the formation of an initiation complex containing 3–4 pairs of bases, and this is sufficient for starting duplex unzipping.
中文翻译:

基于QCM的破裂事件扫描技术是研究DNA双链体解离动力学的一种简单可靠的方法。
破裂事件扫描(REVS)在基于动态力谱的方法中首次应用。使用包含20对寡核苷酸的模型DNA双链体,包括包含单个错配的寡核苷酸,我们证明了可靠确定生物分子复合物解离动力学参数的可能性:屏障位置,解离速率常数和复合物寿命。在这种方法中,DNA双链体的机械解离是根据类似于解压缩的机制发生的。结果表明,该过程是通过克服单个能垒而发生的。在错配位于距QCM表面最远的双链体末端的情况下,观察到结合态和未结合态之间的势垒位置大大降低。
更新日期:2020-08-06
中文翻译:

基于QCM的破裂事件扫描技术是研究DNA双链体解离动力学的一种简单可靠的方法。
破裂事件扫描(REVS)在基于动态力谱的方法中首次应用。使用包含20对寡核苷酸的模型DNA双链体,包括包含单个错配的寡核苷酸,我们证明了可靠确定生物分子复合物解离动力学参数的可能性:屏障位置,解离速率常数和复合物寿命。在这种方法中,DNA双链体的机械解离是根据类似于解压缩的机制发生的。结果表明,该过程是通过克服单个能垒而发生的。在错配位于距QCM表面最远的双链体末端的情况下,观察到结合态和未结合态之间的势垒位置大大降低。











































 京公网安备 11010802027423号
京公网安备 11010802027423号