当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of the Biosynthetic Gene Cluster for the anti-MRSA Lysocins through Gene Cluster Activation Using Strong Promoters of Housekeeping Genes and Production of New Analogs in Lysobacter sp. 3655.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-07-01 , DOI: 10.1021/acssynbio.0c00067 Lingjun Yu 1, 2 , Fengyu Du 2, 3 , Xusheng Chen 2, 4 , Yongbiao Zheng 2, 5 , Martha Morton 2 , Fengquan Liu 1 , Liangcheng Du 2
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-07-01 , DOI: 10.1021/acssynbio.0c00067 Lingjun Yu 1, 2 , Fengyu Du 2, 3 , Xusheng Chen 2, 4 , Yongbiao Zheng 2, 5 , Martha Morton 2 , Fengquan Liu 1 , Liangcheng Du 2
Affiliation
|
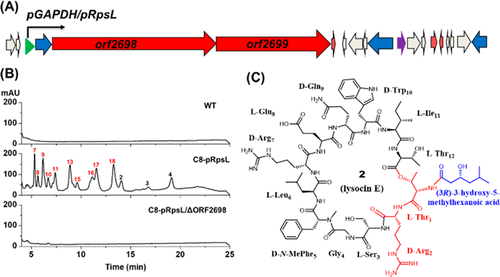
The Gram-negative gliding bacteria Lysobacter represent a new and rich source for bioactive natural products. In an effort to discover new antibiotics, we found a cryptic biosynthetic gene cluster (BGC) in Lysobacter sp. 3655 that shared a high similarity with the putative lysocin BGC identified in silico previously from Lysobacter sp. RH2180-5. Lysocins are cyclic lipodepsipeptides with potent activity against MRSA (methicillin-resistant Staphylococcus aureus) using a novel mode of action, but the lysocin BGC had not been experimentally verified so far. Using an activity-guided screening, we isolated the main antibiotic compound and confirmed it to be lysocin E. However, the putative lysocin BGC was barely transcribed in the wild type, in which lysocins were produced only in specific conditions and in a negligible amount. To activate the putative lysocin BGC, we screened for strongly transcribed housekeeping genes in strain 3655 and found several powerful promoters. Upon engineering the promoters into the BGC, the lysocin gene transcription was significantly enhanced and the lysocin yield was markedly increased. With readily detectable lysocins production in the engineered strains, we showed that lysocin production was abolished in the gene deletion mutant and then restored in the complementary strain, even when grown in conditions that did not support the wild type for lysocin production. Moreover, the engineered strain produced multiple new lysocin congeners. The determination of the lysocin BGC and the Lysobacter promoters will facilitate the ongoing efforts for yield improvement and new antibiotic biosynthesis using synthetic biology strategies.
中文翻译:

通过使用管家基因的强启动子和新的类似物在溶菌属中产生的基因簇激活,通过抗MRSA溶球菌素的生物合成基因簇的鉴定。3655。
革兰氏阴性滑动细菌溶菌菌是生物活性天然产物的新的丰富来源。为了发现新的抗生素,我们在溶杆菌属中发现了一个隐秘的生物合成基因簇(BGC)。3655与假定的溶菌素BGC具有高度相似性,该溶菌素BGC在计算机上已从Lysobacter sp。中鉴定。RH2180-5。溶血球蛋白是对MRSA(耐甲氧西林的金黄色葡萄球菌)具有有效活性的环状脂肽)使用一种新颖的作用方式,但迄今为止尚没有通过实验验证溶菌素BGC。使用活性指导筛选,我们分离出主要的抗生素化合物并确认其为溶菌素E。但是,假定的溶菌素BGC几乎没有在野生型中转录,其中溶菌素仅在特定条件下产生且量可忽略不计。为了激活假定的溶菌素BGC,我们筛选了3655株中强转录的看家基因,并发现了几个强大的启动子。将启动子改造成BGC后,溶菌素基因的转录显着增强,溶菌素的产量明显增加。在工程菌株中可以很容易地检测到溶菌素的产生,我们表明溶菌素的产生在基因缺失突变体中被消除,然后在互补菌株中得以恢复,即使在不支持野生型溶菌素生产的条件下生长。此外,工程菌株产生了多个新的溶菌素同源物。溶血素BGC的测定及血红蛋白的测定。溶菌启动子将促进使用合成生物学策略进行产量提高和新抗生素生物合成的持续努力。
更新日期:2020-08-21
中文翻译:

通过使用管家基因的强启动子和新的类似物在溶菌属中产生的基因簇激活,通过抗MRSA溶球菌素的生物合成基因簇的鉴定。3655。
革兰氏阴性滑动细菌溶菌菌是生物活性天然产物的新的丰富来源。为了发现新的抗生素,我们在溶杆菌属中发现了一个隐秘的生物合成基因簇(BGC)。3655与假定的溶菌素BGC具有高度相似性,该溶菌素BGC在计算机上已从Lysobacter sp。中鉴定。RH2180-5。溶血球蛋白是对MRSA(耐甲氧西林的金黄色葡萄球菌)具有有效活性的环状脂肽)使用一种新颖的作用方式,但迄今为止尚没有通过实验验证溶菌素BGC。使用活性指导筛选,我们分离出主要的抗生素化合物并确认其为溶菌素E。但是,假定的溶菌素BGC几乎没有在野生型中转录,其中溶菌素仅在特定条件下产生且量可忽略不计。为了激活假定的溶菌素BGC,我们筛选了3655株中强转录的看家基因,并发现了几个强大的启动子。将启动子改造成BGC后,溶菌素基因的转录显着增强,溶菌素的产量明显增加。在工程菌株中可以很容易地检测到溶菌素的产生,我们表明溶菌素的产生在基因缺失突变体中被消除,然后在互补菌株中得以恢复,即使在不支持野生型溶菌素生产的条件下生长。此外,工程菌株产生了多个新的溶菌素同源物。溶血素BGC的测定及血红蛋白的测定。溶菌启动子将促进使用合成生物学策略进行产量提高和新抗生素生物合成的持续努力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号