当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
3D variable-density SPARKLING trajectories for high-resolution T2*-weighted magnetic resonance imaging.
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-07-01 , DOI: 10.1002/nbm.4349 Carole Lazarus 1, 2, 3 , Pierre Weiss 4, 5, 6 , Loubna El Gueddari 1, 2, 3 , Franck Mauconduit 7 , Aurélien Massire 1 , Mathilde Ripart 1, 2 , Alexandre Vignaud 1, 2 , Philippe Ciuciu 1, 2, 3
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-07-01 , DOI: 10.1002/nbm.4349 Carole Lazarus 1, 2, 3 , Pierre Weiss 4, 5, 6 , Loubna El Gueddari 1, 2, 3 , Franck Mauconduit 7 , Aurélien Massire 1 , Mathilde Ripart 1, 2 , Alexandre Vignaud 1, 2 , Philippe Ciuciu 1, 2, 3
Affiliation
|
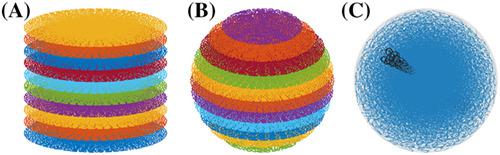
We have recently proposed a new optimization algorithm called SPARKLING (Spreading Projection Algorithm for Rapid K‐space sampLING) to design efficient compressive sampling patterns for magnetic resonance imaging (MRI). This method has a few advantages over conventional non‐Cartesian trajectories such as radial lines or spirals: i) it allows to sample the k‐space along any arbitrary density while the other two are restricted to radial densities and ii) it optimizes the gradient waveforms for a given readout time. Here, we introduce an extension of the SPARKLING method for 3D imaging by considering both stacks‐of‐SPARKLING and fully 3D SPARKLING trajectories. Our method allowed to achieve an isotropic resolution of 600 μ m in just 45 seconds for T2∗‐weighted ex vivo brain imaging at 7 Tesla over a field‐of‐view of 200 × 200 × 140 m m 3. Preliminary in vivo human brain data shows that a stack‐of‐SPARKLING is less subject to off‐resonance artifacts than a stack‐of‐spirals.
中文翻译:

用于高分辨率 T2* 加权磁共振成像的 3D 可变密度 SPARKLING 轨迹。
我们最近提出了一种称为 SPARKLING(快速 K 空间采样的扩展投影算法)的新优化算法,以设计用于磁共振成像 (MRI) 的高效压缩采样模式。与传统的非笛卡尔轨迹(例如径向线或螺旋线)相比,这种方法具有一些优点:i)它允许沿任意密度对 k 空间进行采样,而其他两种方法仅限于径向密度;ii)它优化了梯度波形对于给定的读出时间。在这里,我们通过考虑堆栈的 SPARKLING 和完全 3D SPARKLING 轨迹来介绍 SPARKLING 方法的扩展,用于 3D 成像。我们的方法允许实现为600的各向同性分辨率μ米在刚刚45秒对于T 2 *200 × 200 × 140 m m 3视场内 7 特斯拉的加权离体脑成像。初步的体内人脑数据显示,SPARKLING 堆栈比螺旋堆栈更不容易受到偏共振伪影的影响。
更新日期:2020-08-04
中文翻译:

用于高分辨率 T2* 加权磁共振成像的 3D 可变密度 SPARKLING 轨迹。
我们最近提出了一种称为 SPARKLING(快速 K 空间采样的扩展投影算法)的新优化算法,以设计用于磁共振成像 (MRI) 的高效压缩采样模式。与传统的非笛卡尔轨迹(例如径向线或螺旋线)相比,这种方法具有一些优点:i)它允许沿任意密度对 k 空间进行采样,而其他两种方法仅限于径向密度;ii)它优化了梯度波形对于给定的读出时间。在这里,我们通过考虑堆栈的 SPARKLING 和完全 3D SPARKLING 轨迹来介绍 SPARKLING 方法的扩展,用于 3D 成像。我们的方法允许实现为600的各向同性分辨率μ米在刚刚45秒对于T 2 *200 × 200 × 140 m m 3视场内 7 特斯拉的加权离体脑成像。初步的体内人脑数据显示,SPARKLING 堆栈比螺旋堆栈更不容易受到偏共振伪影的影响。



























 京公网安备 11010802027423号
京公网安备 11010802027423号