iScience ( IF 4.6 ) Pub Date : 2020-07-01 , DOI: 10.1016/j.isci.2020.101321 Stephen Cole 1 , Sudhakaran Prabakaran 1
|
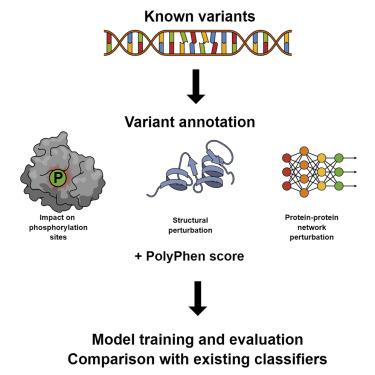
Phosphorylation sites often have key regulatory functions and are central to many cellular signaling pathways, so mutations that modify them have the potential to contribute to pathological states such as cancer. Although many classifiers exist for prioritization of coding genomic variants, to our knowledge none of them explicitly account for the alteration or creation of kinase recognition motifs that alter protein structure, function, regulation of activity, and interaction networks through modifying the pattern of phosphorylation. We present a novel computational pipeline that uses a random forest classifier to predict the pathogenicity of a variant, according to its direct or indirect effect on local phosphorylation sites and the predicted functional impact of perturbing a phosphorylation event. We call this classifier PhosphoEffect and find that it compares favorably and with increased accuracy to the existing classifier PolyPhen 2.2.2 when tested on a dataset of known variants enriched for phosphorylation sites and their neighbors.
中文翻译:

PhosphoEffect:通过对激酶识别基序的影响来优先考虑磷酸化位点上或邻近磷酸化位点的变体。
磷酸化位点通常具有关键的调节功能,并且是许多细胞信号传导途径的核心,因此修饰它们的突变有可能导致癌症等病理状态。尽管存在许多分类器用于编码基因组变体的优先级,但据我们所知,它们都没有明确解释激酶识别基序的改变或创建,这些激酶识别基序通过修改磷酸化模式来改变蛋白质结构、功能、活性调节和相互作用网络。我们提出了一种新颖的计算管道,它使用随机森林分类器根据变异对局部磷酸化位点的直接或间接影响以及扰乱磷酸化事件的预测功能影响来预测变异的致病性。我们将此分类器称为 PhosphoEffect,并发现当在富含磷酸化位点及其邻近位点的已知变体数据集上进行测试时,它与现有分类器 PolyPhen 2.2.2 相比具有更好的性能和更高的准确性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号