当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Release Factor Inhibiting Antimicrobial Peptides Improve Nonstandard Amino Acid Incorporation in Wild-type Bacterial Cells.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-06-30 , DOI: 10.1021/acschembio.0c00055 Erkin Kuru 1, 2 , Rosa-Maria Määttälä 1, 3 , Karen Noguera 1 , Devon A Stork 1 , Kamesh Narasimhan 1 , Jonathan Rittichier 1, 2 , Daniel Wiegand 1, 2 , George M Church 1, 2
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-06-30 , DOI: 10.1021/acschembio.0c00055 Erkin Kuru 1, 2 , Rosa-Maria Määttälä 1, 3 , Karen Noguera 1 , Devon A Stork 1 , Kamesh Narasimhan 1 , Jonathan Rittichier 1, 2 , Daniel Wiegand 1, 2 , George M Church 1, 2
Affiliation
|
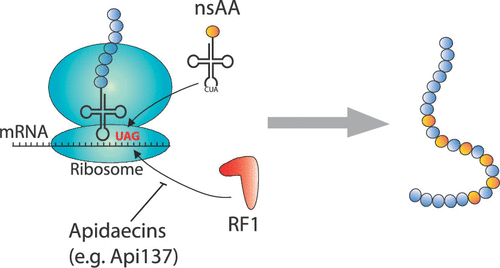
We report a tunable chemical genetics approach for enhancing genetic code expansion in different wild-type bacterial strains that employ apidaecin-like, antimicrobial peptides observed to temporarily sequester and thereby inhibit Release Factor 1 (RF1). In a concentration-dependent matter, these peptides granted a conditional lambda phage resistance to a recoded Escherichia coli strain with nonessential RF1 activity and promoted multisite nonstandard amino acid (nsAA) incorporation at in-frame amber stop codons in vivo and in vitro. When exogenously added, the peptides stimulated specific nsAA incorporation in a variety of sensitive, wild-type (RF1+) strains, including Agrobacterium tumefaciens, a species in which nsAA incorporation has not been previously reported. Improvement in nsAA incorporation was typically 2–15-fold in E. coli BL21, MG1655, and DH10B strains and A. tumefaciens with the >20-fold improvement observed in probiotic E. coli Nissle 1917. In-cell expression of these peptides promoted multisite nsAA incorporation in transcripts with up to 6 amber codons, with a >35-fold increase in BL21 showing moderate toxicity. Leveraging this RF1 sensitivity allowed multiplexed partial recoding of MG1655 and DH10B that rapidly resulted in resistant strains that showed an additional approximately twofold boost to nsAA incorporation independent of the peptide. Finally, in-cell expression of an apidaecin-like peptide library allowed the discovery of new peptide variants with reduced toxicity that still improved multisite nsAA incorporation >25-fold. In parallel to genetic reprogramming efforts, these new approaches can facilitate genetic code expansion technologies in a variety of wild-type bacterial strains.
中文翻译:

释放因子抑制抗菌肽可改善野生型细菌细胞中非标准氨基酸的掺入。
我们报告了一种可调谐的化学遗传学方法,用于增强在不同的野生型细菌菌株中的遗传密码扩展,这些菌株使用了能暂时隔离并从而抑制释放因子1(RF1)的阿迪霉素样抗微生物肽。在浓度依赖性的情况下,这些肽对具有非必需RF1活性的经重新编码的大肠杆菌菌株赋予了条件性λ噬菌体抗性,并在体内和体外在框内琥珀终止密码子处促进了多位非标准氨基酸(nsAA)掺入。当外源添加时,这些肽可刺激特定的nsAA掺入多种敏感的野生型(RF1 +)菌株中,包括根癌农杆菌,以前没有报道过nsAA掺入的物种。在大肠杆菌BL21,MG1655和DH10B菌株和根癌农杆菌中,nsAA掺入的改善通常为2–15倍,而在益生菌大肠杆菌中观察到的> 20倍改善Nissle1917。这些肽在细胞内的表达促进了多位点nsAA掺入具有多达6个琥珀色密码子的转录物中,而BL21的增加> 35倍,显示出中等毒性。利用此RF1敏感性,可以对MG1655和DH10B进行多重部分编码,从而迅速产生耐药菌株,这些菌株对nsAA的掺入表现出额外的大约两倍的增强,而与肽无关。最终,在细胞中表达一种类似黄霉素的肽文库可以发现毒性降低的新肽变体,该变体仍使多位nsAA掺入提高了25倍以上。与基因重编程工作并行,这些新方法可以促进多种野生型细菌菌株中的遗传密码扩展技术。
更新日期:2020-07-17
中文翻译:

释放因子抑制抗菌肽可改善野生型细菌细胞中非标准氨基酸的掺入。
我们报告了一种可调谐的化学遗传学方法,用于增强在不同的野生型细菌菌株中的遗传密码扩展,这些菌株使用了能暂时隔离并从而抑制释放因子1(RF1)的阿迪霉素样抗微生物肽。在浓度依赖性的情况下,这些肽对具有非必需RF1活性的经重新编码的大肠杆菌菌株赋予了条件性λ噬菌体抗性,并在体内和体外在框内琥珀终止密码子处促进了多位非标准氨基酸(nsAA)掺入。当外源添加时,这些肽可刺激特定的nsAA掺入多种敏感的野生型(RF1 +)菌株中,包括根癌农杆菌,以前没有报道过nsAA掺入的物种。在大肠杆菌BL21,MG1655和DH10B菌株和根癌农杆菌中,nsAA掺入的改善通常为2–15倍,而在益生菌大肠杆菌中观察到的> 20倍改善Nissle1917。这些肽在细胞内的表达促进了多位点nsAA掺入具有多达6个琥珀色密码子的转录物中,而BL21的增加> 35倍,显示出中等毒性。利用此RF1敏感性,可以对MG1655和DH10B进行多重部分编码,从而迅速产生耐药菌株,这些菌株对nsAA的掺入表现出额外的大约两倍的增强,而与肽无关。最终,在细胞中表达一种类似黄霉素的肽文库可以发现毒性降低的新肽变体,该变体仍使多位nsAA掺入提高了25倍以上。与基因重编程工作并行,这些新方法可以促进多种野生型细菌菌株中的遗传密码扩展技术。









































 京公网安备 11010802027423号
京公网安备 11010802027423号