Journal of Proteomics ( IF 2.8 ) Pub Date : 2020-06-30 , DOI: 10.1016/j.jprot.2020.103887 Karim Hayoun 1 , Jean-Charles Gaillard 1 , Olivier Pible 1 , Béatrice Alpha-Bazin 1 , Jean Armengaud 1
|
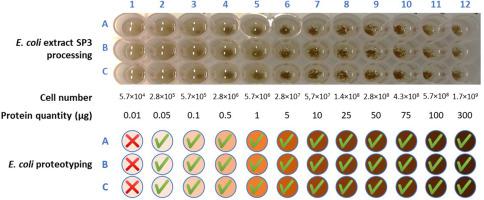
Tandem mass spectrometry-based proteotyping of microorganisms presents several advantages over whole-cell MALDI-TOF mass spectrometry: because a larger number of signals are recorded with better accuracy and precision, the approach allows for the identification of microorganisms at more resolved taxonomic levels, and can easily manage complex samples. Additionally, the use of SP3 paramagnetic beads for cell lysis and protein cleanup simplifies sample preparation for proteotyping. Based on these features, we have developed and tested a 96-well plate platform for high-throughput proteotyping of a large variety of bacteria. We evaluated the performance of the platform in terms of bacterial load and found no cross-contamination between wells. Likewise, phylopeptidomics analysis revealed no alteration in the relative quantifications of microorganisms. Finally, we applied this new format for rapid proteotyping of a large set of dental isolates using double-barrel chromatography coupled to tandem mass spectrometry, which maximizes the number of spectra per unit of time. The procedure allowed us to establish whether these isolates were pure strains or mixtures of strains and to identify the microorganisms at the most resolved taxonomic level.
Significance
The rapid and accurate identification of microorganisms is a clinical priority in medical diagnostics; however, specimens containing mixtures of microorganisms are difficult to analyze and the procedure is time-consuming. Tandem mass spectrometry proteotyping allows the fast identification of complex mixtures of microorganisms, known or unknown, and can also establish the biomass ratio of each component. We describe here a new workflow for preparing microbial samples in a 96-well-plate format for tandem mass spectrometry proteotyping and document its advantages and limitations. We demonstrate that this new format coupled to a highly efficient double-barrel LC-MS/MS system allows proteotyping of 96 isolates in 55 h.
中文翻译:

基于微孔板顺磁珠和系统肽组学的双桶色谱-串联质谱技术对细菌分离物进行高通量蛋白质分型。
与全细胞MALDI-TOF质谱相比,基于串联质谱的微生物蛋白鉴定具有多个优势:由于以更高的准确性和精度记录了大量信号,因此该方法可以在更分解的分类学水平上鉴定微生物,并且可以轻松管理复杂的样本。此外,使用SP3顺磁珠进行细胞裂解和蛋白质纯化可简化样品制备过程,以进行蛋白质分型。基于这些功能,我们开发并测试了96孔板平台,可用于多种细菌的高通量蛋白质分型。我们根据细菌载量评估了平台的性能,发现孔之间没有交叉污染。同样,系统肽组学分析显示微生物的相对定量没有变化。最后,我们将这种新格式应用于使用双管色谱与串联质谱联用的大量牙科分离株的快速蛋白原型化,从而使单位时间内的光谱数最大化。该程序使我们能够确定这些分离株是纯菌株还是菌株混合物,并能以最有效的分类学水平鉴定微生物。
意义
快速准确地鉴定微生物是医学诊断中的临床优先事项。但是,含有微生物混合物的标本很难分析,而且过程耗时。串联质谱分析技术可以快速鉴定已知或未知的复杂微生物混合物,还可以确定每种组分的生物量比率。我们在这里描述了一种新的工作流程,该工作流程用于制备96孔板格式的微生物样品,用于串联质谱法原型设计,并记录其优势和局限性。我们证明,这种新形式与高效的双桶LC-MS / MS系统耦合,可以在55小时内对96个分离株进行蛋白分型。











































 京公网安备 11010802027423号
京公网安备 11010802027423号