Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-06-29 , DOI: 10.1016/j.jbi.2020.103484 Zhili Zhao 1 , Jian Qin 1 , Zhuoyue Gou 1 , Yanan Zhang 1 , Yi Yang 1
|
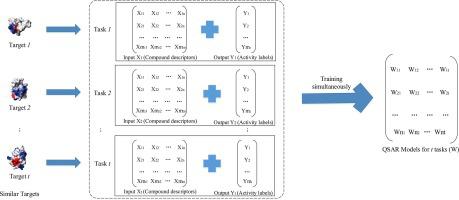
The computational drug discovery methods can find potential drug-target interactions more efficiently and have been widely studied over past few decades. Such methods explore the relationship between the structural properties of compounds and their biological activity with the assumption that similar compounds tend to share similar biological targets and vice versa. However, traditional Quantitative Structure - Activity Relationship (QSAR) methods often do not have desired accuracy due to insufficient data of compound activity. In this paper, we focus on building Multi-Task Learning (MTL)-based QSAR models by considering multiple similar biological targets together and make shared information transfer across from one task to another, thereby improving not only the learning efficiency, but also the prediction accuracy. This paper selects 6 assay groups with similar biological targets from PubChem and builds their QSAR models with MTL simultaneously. According to the experiment results, our MTL-based QSAR models have better performance over traditional prominent machine learning algorithms and the improvements are even more obvious when other baseline models have low accuracy. The superiority of our models is also proved by Student’s t-test with level of significance 5%. Moreover, this paper also explores three different assumptions on the underlying pattern in the dataset and finds that the joint feature MTL models further improve the performance of the QSAR models and are more suitable for building QSAR models for multiple similar biological targets.
中文翻译:

用于预测活性化合物的多任务学习模型。
计算药物发现方法可以更有效地发现潜在的药物-靶标相互作用,并且在过去的几十年中已得到广泛研究。假设相似的化合物倾向于共享相似的生物靶标,反之亦然,这样的方法探索了化合物的结构特性与其生物学活性之间的关系。但是,由于化合物活性的数据不足,传统的定量结构-活性关系(QSAR)方法通常没有所需的准确性。在本文中,我们专注于通过同时考虑多个相似的生物学目标来构建基于多任务学习(MTL)的QSAR模型,并使共享信息从一项任务转移到另一项任务,从而不仅提高了学习效率,而且还提高了预测能力准确性。本文从PubChem中选择了具有相似生物学目标的6个检测组,并同时建立了具有MTL的QSAR模型。根据实验结果,我们的基于MTL的QSAR模型比传统的著名机器学习算法具有更好的性能,而当其他基线模型的准确性较低时,改进甚至更加明显。学生模型也证明了我们模型的优越性t检验的显着性水平为5%。此外,本文还探讨了数据集中基础模式的三个不同假设,并发现联合特征MTL模型进一步提高了QSAR模型的性能,更适合于为多个相似的生物目标建立QSAR模型。



























 京公网安备 11010802027423号
京公网安备 11010802027423号