Letters in Drug Design & Discovery ( IF 1.2 ) Pub Date : 2020-05-31 , DOI: 10.2174/1570180816666190712095907 Uttam Ashok More 1 , Sameera Patel 1 , Vidhi Rahevar 1 , Malleshappa Ningappa Noolvi 1 , Tejraj M. Aminabhavi 1 , Shrinivas D. Joshi 2
|
Background: Alzheimer’s disease (AD) is increasingly being recognized as one of the lethal diseases in older people. Acetylcholinesterase (AChE) has proven to be the most promising target in AD, used for designing drugs against AD.
Methods: In silico studies, 2D- or 3D-QSAR like hologram QSAR (HQSAR), Topomer comparative molecular field analysis (Topomer CoMFA), comparative molecular field analysis (CoMFA), and comparative molecular similarity indices analysis (CoMSIA) methods were used to generate QSAR models for acetylcholinesterase inhibitors.
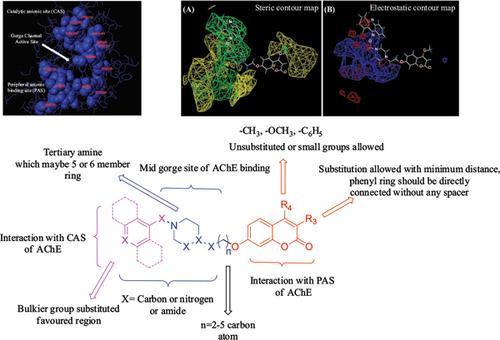
Results: Acetylcholinesterase inhibitors used for the present study contain a series of 7- hydroxycoumarin derivatives connected by piperidine, piperazine, tacrine, triazole, or benzyl fragments through alkyl or amide spacer training set compounds were used to generate best model with a HQSAR q2 value of 0.916 and r2 value of 0.940; a Topomer CoMFA q2 value of 0.907 and r2 value of 0.959, CoMFA q2 value of 0.880 and r2 value of 0.960; and a CoMSIA q2 value of 0.865 and r2 value of 0.941. In addition, contour plots obtained from QSAR models suggested the significant regions that influenced the AChE inhibitory activity.
Conclusion: In light of these results, this study provides knowledge about the structural requirements for the development of more active acetylcholinesterase inhibitors. In addition, the predicted ADME profile helps us to find CNS active molecules, the obtained prediction compared with well-known AChE inhibitors viz., ensaculin, tacrine, galantamine, rivastigmine, and donepezil. Based on the knowledge obtained from these studies, the hybridization approach is one of the best ways to find lead compounds and these findings can be useful in the treatment of Alzheimer's disease.
中文翻译:

在Silico ADME和QSAR中研究了一组香豆素衍生物作为抗阿尔茨海默氏病的乙酰胆碱酯酶抑制剂:CoMFA,CoMSIA,Topomer CoMFA和HQSAR
背景:阿尔茨海默氏病(AD)越来越被认为是老年人的致死性疾病之一。乙酰胆碱酯酶(AChE)已被证明是AD中最有希望的靶标,用于设计抗AD药物。
方法:在计算机模拟研究中,使用2D-或3D-QSAR全息图QSAR(HQSAR),拓扑异构体比较分子场分析(Topomer CoMFA),比较分子场分析(CoMFA)和比较分子相似性指标分析(CoMSIA)方法生成乙酰胆碱酯酶抑制剂的QSAR模型。
结果:用于本研究的乙酰胆碱酯酶抑制剂包含一系列7-羟基香豆素衍生物,它们通过烷基或酰胺间隔基训练集化合物与哌啶,哌嗪,他克林,三唑或苄基片段连接,用于生成HQSAR q2值为0.916和r2值为0.940;拓扑CoMFA q2值为0.907,r2值为0.959,CoMFA q2值为0.880,r2值为0.960;CoMSIA q2值为0.865,r2值为0.941。此外,从QSAR模型获得的等高线图表明了影响AChE抑制活性的重要区域。
结论:根据这些结果,本研究提供了有关开发更具活性的乙酰胆碱酯酶抑制剂的结构要求的知识。此外,预测的ADME谱有助于我们找到CNS活性分子,与知名的AChE抑制剂(即,依斯考林,他克林,加兰他敏,卡巴拉汀和多奈哌齐)相比,获得的预测值更好。根据从这些研究中获得的知识,杂交方法是寻找先导化合物的最佳方法之一,这些发现可用于治疗阿尔茨海默氏病。











































 京公网安备 11010802027423号
京公网安备 11010802027423号