Current Proteomics ( IF 0.5 ) Pub Date : 2020-07-31 , DOI: 10.2174/1570164616666190306152318 Chang Xu 1 , Limin Jiang 1 , Zehua Zhang 1 , Xuyao Yu 2 , Renhai Chen 1 , Junhai Xu 1
|
Background: Protein-Protein Interactions (PPIs) play a key role in various biological processes. Many methods have been developed to predict protein-protein interactions and protein interaction networks. However, many existing applications are limited, because of relying on a large number of homology proteins and interaction marks.
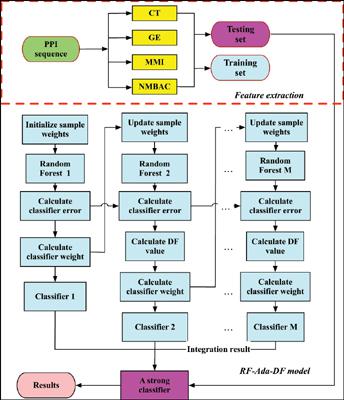
Methods: In this paper, we propose a novel integrated learning approach (RF-Ada-DF) with the sequence-based feature representation, for identifying protein-protein interactions. Our method firstly constructs a sequence-based feature vector to represent each pair of proteins, via Multivariate Mutual Information (MMI) and Normalized Moreau-Broto Autocorrelation (NMBAC). Then, we feed the 638- dimentional features into an integrated learning model for judging interaction pairs and non-interaction pairs. Furthermore, this integrated model embeds Random Forest in AdaBoost framework and turns weak classifiers into a single strong classifier. Meanwhile, we also employ double fault detection in order to suppress over-adaptation during the training process.
Results: To evaluate the performance of our method, we conduct several comprehensive tests for PPIs prediction. On the H. pylori dataset, our method achieves 88.16% accuracy and 87.68% sensitivity, the accuracy of our method is increased by 0.57%. On the S. cerevisiae dataset, our method achieves 95.77% accuracy and 93.36% sensitivity, the accuracy of our method is increased by 0.76%. On the Human dataset, our method achieves 98.16% accuracy and 96.80% sensitivity, the accuracy of our method is increased by 0.6%. Experiments show that our method achieves better results than other outstanding methods for sequence-based PPIs prediction. The datasets and codes are available at https://github.com/guofei-tju/RF-Ada-DF.git.
中文翻译:

蛋白质-蛋白质相互作用的综合预测方法
背景:蛋白质-蛋白质相互作用(PPI)在各种生物过程中起着关键作用。已经开发出许多方法来预测蛋白质-蛋白质相互作用和蛋白质相互作用网络。然而,由于依赖于大量的同源蛋白和相互作用标记,许多现有的应用受到限制。
方法:在本文中,我们提出了一种新颖的基于序列特征表示的集成学习方法(RF-Ada-DF),用于识别蛋白质-蛋白质相互作用。我们的方法首先通过多元互信息(MMI)和标准化的Moreau-Broto自相关(NMBAC)构建了一个基于序列的特征向量来表示每对蛋白质。然后,我们将638维特征馈入一个集成的学习模型中,以判断交互对和非交互对。此外,该集成模型将随机森林嵌入AdaBoost框架中,并将弱分类器转变为单个强分类器。同时,我们还采用双重故障检测以抑制训练过程中的过度适应。
结果:为了评估我们方法的性能,我们对PPI进行了一些综合测试。在幽门螺杆菌数据集上,我们的方法达到了88.16%的准确度和87.68%的灵敏度,我们的方法的准确度提高了0.57%。在酿酒酵母数据集上,我们的方法达到了95.77%的准确度和93.36%的灵敏度,我们的方法的准确度提高了0.76%。在人类数据集上,我们的方法达到98.16%的准确性和96.80%的灵敏度,我们的方法的准确性提高了0.6%。实验表明,与其他出色的基于序列的PPI预测方法相比,我们的方法取得了更好的结果。数据集和代码位于https://github.com/guofei-tju/RF-Ada-DF.git。











































 京公网安备 11010802027423号
京公网安备 11010802027423号