Current Proteomics ( IF 0.5 ) Pub Date : 2020-07-31 , DOI: 10.2174/1570164616666190417100509 Yijie Ding 1 , Feng Chen 1 , Xiaoyi Guo 2 , Jijun Tang 3 , Hongjie Wu 1
|
Background: The DNA-binding proteins is an important process in multiple biomolecular functions. However, the tradition experimental methods for DNA-binding proteins identification are still time consuming and extremely expensive.
Objective: In past several years, various computational methods have been developed to detect DNAbinding proteins. However, most of them do not integrate multiple information.
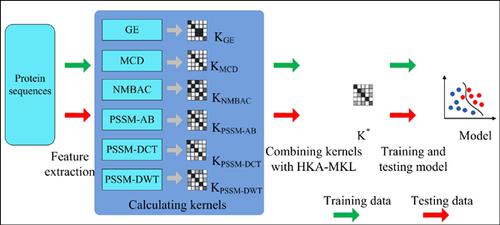
Methods: In this study, we propose a novel computational method to predict DNA-binding proteins by two steps Multiple Kernel Support Vector Machine (MK-SVM) and sequence information. Firstly, we extract several feature and construct multiple kernels. Then, multiple kernels are linear combined by Multiple Kernel Learning (MKL). At last, a final SVM model, constructed by combined kernel, is built to predict DNA-binding proteins.
Results: The proposed method is tested on two benchmark data sets. Compared with other existing method, our approach is comparable, even better than other methods on some data sets.
Conclusion: We can conclude that MK-SVM is more suitable than common SVM, as the classifier for DNA-binding proteins identification.
中文翻译:

多核支持向量机和序列信息鉴定DNA结合蛋白
背景:DNA结合蛋白是多种生物分子功能的重要过程。然而,用于DNA结合蛋白鉴定的传统实验方法仍然是耗时且极其昂贵的。
目的:在过去的几年中,已经开发出各种计算方法来检测DNA结合蛋白。但是,它们中的大多数不集成多个信息。
方法:在这项研究中,我们提出了一种通过两步多核支持向量机(MK-SVM)和序列信息预测DNA结合蛋白的新颖计算方法。首先,我们提取几个特征并构造多个内核。然后,通过多核学习(MKL)将多个核线性组合。最后,通过组合核构建最终的SVM模型,以预测DNA结合蛋白。
结果:所提出的方法在两个基准数据集上进行了测试。与其他现有方法相比,我们的方法具有可比性,甚至在某些数据集上也优于其他方法。
结论:我们可以得出结论,MK-SVM比普通SVM更适合作为DNA结合蛋白鉴定的分类器。











































 京公网安备 11010802027423号
京公网安备 11010802027423号