当前位置:
X-MOL 学术
›
STEM CELLS
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The epitranscriptome landscape of small noncoding RNAs in stem cells
STEM CELLS ( IF 4.0 ) Pub Date : 2020-06-29 , DOI: 10.1002/stem.3233 James M W R McElhinney 1 , Ayesha Hasan 1 , Abdulrahim A Sajini 1
STEM CELLS ( IF 4.0 ) Pub Date : 2020-06-29 , DOI: 10.1002/stem.3233 James M W R McElhinney 1 , Ayesha Hasan 1 , Abdulrahim A Sajini 1
Affiliation
|
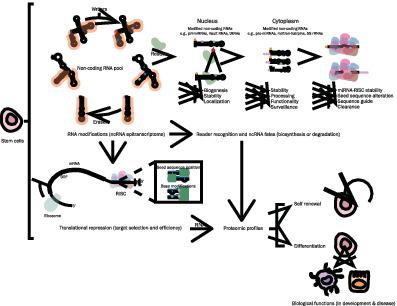
Stem cells (SCs) are unique cells that have an inherent ability to self‐renew or differentiate. Both fate decisions are strongly regulated at the molecular level via intricate signaling pathways. The regulation of signaling networks promoting self‐renewal or differentiation was thought to be largely governed by the action of transcription factors. However, small noncoding RNAs (ncRNAs), such as vault RNAs, and their post‐transcriptional modifications (the epitranscriptome) have emerged as additional regulatory layers with essential roles in SC fate decisions. RNA post‐transcriptional modifications often modulate RNA stability, splicing, processing, recognition, and translation. Furthermore, modifications on small ncRNAs allow for dual regulation of RNA activity, at both the level of biogenesis and RNA‐mediated actions. RNA post‐transcriptional modifications act through structural alterations and specialized RNA‐binding proteins (RBPs) called writers, readers, and erasers. It is through SC‐context RBPs that the epitranscriptome coordinates specific functional roles. Small ncRNA post‐transcriptional modifications are today exploited by different mechanisms to facilitate SC translational studies. One mechanism readily being studied is identifying how SC‐specific RBPs of small ncRNAs regulate fate decisions. Another common practice of using the epitranscriptome for regenerative applications is using naturally occurring post‐transcriptional modifications on synthetic RNA to generate induced pluripotent SCs. Here, we review exciting insights into how small ncRNA post‐transcriptional modifications control SC fate decisions in development and disease. We hope, by illustrating how essential the epitranscriptome and their associated proteome are in SCs, they would be considered as novel tools to propagate SCs for regenerative medicine.
中文翻译:

干细胞中小非编码 RNA 的表观转录组景观
干细胞 (SC) 是独特的细胞,具有自我更新或分化的内在能力。这两种命运决定都通过复杂的信号通路在分子水平上受到强烈调控。促进自我更新或分化的信号网络的调节被认为在很大程度上受转录因子的作用控制。然而,小的非编码 RNA (ncRNA),例如 vault RNA,及其转录后修饰(表观转录组)已成为额外的调控层,在 SC 命运决定中发挥重要作用。RNA 转录后修饰通常调节 RNA 的稳定性、剪接、加工、识别和翻译。此外,对小 ncRNA 的修饰允许在生物发生和 RNA 介导的作用水平上对 RNA 活性进行双重调节。RNA 转录后修饰通过结构改变和称为写入器、读取器和擦除器的特殊 RNA 结合蛋白 (RBP) 起作用。正是通过 SC 上下文 RBP,表观转录组协调特定的功能作用。小 ncRNA 转录后修饰如今被不同的机制利用来促进 SC 翻译研究。一种正在研究的机制是确定小 ncRNA 的 SC 特异性 RBP 如何调节命运决定。使用表观转录组进行再生应用的另一种常见做法是对合成 RNA 使用自然发生的转录后修饰来产生诱导的多能干细胞。在这里,我们回顾了关于小 ncRNA 转录后修饰如何控制发育和疾病中 SC 命运决定的令人兴奋的见解。我们希望,
更新日期:2020-06-29
中文翻译:

干细胞中小非编码 RNA 的表观转录组景观
干细胞 (SC) 是独特的细胞,具有自我更新或分化的内在能力。这两种命运决定都通过复杂的信号通路在分子水平上受到强烈调控。促进自我更新或分化的信号网络的调节被认为在很大程度上受转录因子的作用控制。然而,小的非编码 RNA (ncRNA),例如 vault RNA,及其转录后修饰(表观转录组)已成为额外的调控层,在 SC 命运决定中发挥重要作用。RNA 转录后修饰通常调节 RNA 的稳定性、剪接、加工、识别和翻译。此外,对小 ncRNA 的修饰允许在生物发生和 RNA 介导的作用水平上对 RNA 活性进行双重调节。RNA 转录后修饰通过结构改变和称为写入器、读取器和擦除器的特殊 RNA 结合蛋白 (RBP) 起作用。正是通过 SC 上下文 RBP,表观转录组协调特定的功能作用。小 ncRNA 转录后修饰如今被不同的机制利用来促进 SC 翻译研究。一种正在研究的机制是确定小 ncRNA 的 SC 特异性 RBP 如何调节命运决定。使用表观转录组进行再生应用的另一种常见做法是对合成 RNA 使用自然发生的转录后修饰来产生诱导的多能干细胞。在这里,我们回顾了关于小 ncRNA 转录后修饰如何控制发育和疾病中 SC 命运决定的令人兴奋的见解。我们希望,











































 京公网安备 11010802027423号
京公网安备 11010802027423号