当前位置:
X-MOL 学术
›
Immunol. Cell Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Time-resolved mRNA and miRNA expression profiling reveals crucial coregulation of molecular pathways involved in epithelial-pneumococcal interactions.
Immunology and Cell Biology ( IF 3.2 ) Pub Date : 2020-06-27 , DOI: 10.1111/imcb.12371 Haiyan Li 1 , Li Lin 1 , Lei Chong 1 , Shuge Gu 1 , Shunhang Wen 1 , Gang Yu 1 , Xiaoguang Hu 1 , Lin Dong 1 , Hailin Zhang 1 , Changchong Li 1
Immunology and Cell Biology ( IF 3.2 ) Pub Date : 2020-06-27 , DOI: 10.1111/imcb.12371 Haiyan Li 1 , Li Lin 1 , Lei Chong 1 , Shuge Gu 1 , Shunhang Wen 1 , Gang Yu 1 , Xiaoguang Hu 1 , Lin Dong 1 , Hailin Zhang 1 , Changchong Li 1
Affiliation
|
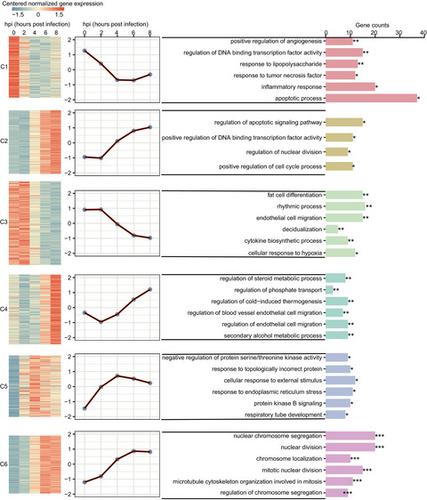
Streptococcus pneumoniae is a major causative agent of pneumonia worldwide and its complex interaction with the lung epithelium has not been thoroughly characterized. In this study, we exploited both RNA‐sequencing and microRNA (miRNA)‐sequencing approaches to monitor the transcriptional changes in human lung alveolar epithelial cells infected by S. pneumoniae in a time‐resolved manner. A total of 1330 differentially expressed (DE) genes and 45 DE miRNAs were identified in all comparisons during the infection process. Clustering analysis showed that all DE genes were grouped into six clusters, several of which were primarily involved in inflammatory or immune responses. In addition, target gene enrichment analyses identified 11 transcription factors that were predicted to link at least one of four clusters, revealing transcriptional coregulation of multiple processes or pathways by common transcription factors. Notably, pharmacological treatment suggested that phosphorylation of p65 is important for optimal transcriptional regulation of target genes in epithelial cells exposed to pathogens. Furthermore, network‐based clustering analysis separated the DE genes negatively regulated by DE miRNAs into two functional modules (M1 and M2), with an enrichment in immune responses and apoptotic signaling pathways for M1. Integrated network analyses of potential regulatory interactions in M1 revealed that multiple DE genes related to immunity and apoptosis were regulated by multiple miRNAs, indicating the coordinated regulation of multiple genes by multiple miRNAs. In conclusion, time‐series expression profiling of messenger RNA and miRNA provides a wealth of information for global transcriptional changes, and offers comprehensive insight into the molecular mechanisms underlying host–pathogen interactions.
中文翻译:

时间分辨的 mRNA 和 miRNA 表达谱揭示了参与上皮-肺炎球菌相互作用的分子途径的关键协同调节。
肺炎链球菌是世界范围内肺炎的主要病原体,其与肺上皮的复杂相互作用尚未得到彻底表征。在这项研究中,我们利用 RNA 测序和 microRNA (miRNA) 测序方法来监测被肺炎链球菌感染的人肺泡上皮细胞的转录变化以时间解析的方式。在感染过程中的所有比较中,共鉴定出 1330 个差异表达 (DE) 基因和 45 个 DE miRNA。聚类分析显示所有 DE 基因分为六个簇,其中几个主要参与炎症或免疫反应。此外,目标基因富集分析确定了 11 个转录因子,这些因子预计将连接四个簇中的至少一个,揭示了常见转录因子对多个过程或途径的转录共调控。值得注意的是,药物治疗表明 p65 的磷酸化对于暴露于病原体的上皮细胞中靶基因的最佳转录调控很重要。此外,基于网络的聚类分析将受 DE miRNA 负调控的 DE 基因分为两个功能模块(M1 和 M2),其中富集了 M1 的免疫反应和凋亡信号通路。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。
更新日期:2020-06-27
中文翻译:

时间分辨的 mRNA 和 miRNA 表达谱揭示了参与上皮-肺炎球菌相互作用的分子途径的关键协同调节。
肺炎链球菌是世界范围内肺炎的主要病原体,其与肺上皮的复杂相互作用尚未得到彻底表征。在这项研究中,我们利用 RNA 测序和 microRNA (miRNA) 测序方法来监测被肺炎链球菌感染的人肺泡上皮细胞的转录变化以时间解析的方式。在感染过程中的所有比较中,共鉴定出 1330 个差异表达 (DE) 基因和 45 个 DE miRNA。聚类分析显示所有 DE 基因分为六个簇,其中几个主要参与炎症或免疫反应。此外,目标基因富集分析确定了 11 个转录因子,这些因子预计将连接四个簇中的至少一个,揭示了常见转录因子对多个过程或途径的转录共调控。值得注意的是,药物治疗表明 p65 的磷酸化对于暴露于病原体的上皮细胞中靶基因的最佳转录调控很重要。此外,基于网络的聚类分析将受 DE miRNA 负调控的 DE 基因分为两个功能模块(M1 和 M2),其中富集了 M1 的免疫反应和凋亡信号通路。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。M1 中潜在调控相互作用的综合网络分析表明,多个与免疫和凋亡相关的 DE 基因受多个 miRNA 的调控,表明多个 miRNA 对多个基因的协同调控。总之,信使 RNA 和 miRNA 的时间序列表达谱为全球转录变化提供了丰富的信息,并提供了对宿主 - 病原体相互作用的分子机制的全面了解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号