当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mobilization of pdif modules in Acinetobacter: A novel mechanism for antibiotic resistance gene shuffling?
Molecular Microbiology ( IF 2.6 ) Pub Date : 2020-06-28 , DOI: 10.1111/mmi.14563 Phillip Balalovski 1 , Ian Grainge 1
Molecular Microbiology ( IF 2.6 ) Pub Date : 2020-06-28 , DOI: 10.1111/mmi.14563 Phillip Balalovski 1 , Ian Grainge 1
Affiliation
|
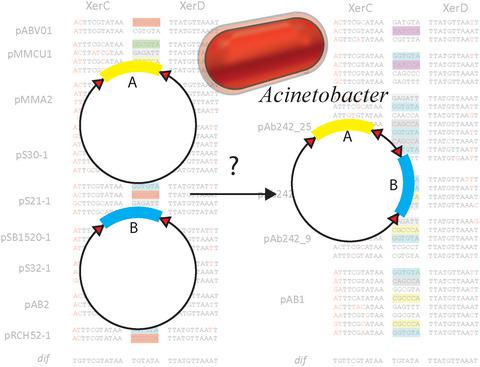
XerCD‐dif site‐specific recombination is a well characterized system, found in most bacteria and archaea. Its role is resolution of chromosomal dimers that arise from homologous recombination. Xer‐mediated recombination is also used by several plasmids for multimer resolution to enhance stability and by some phage for integration into the chromosome. In the past decade, it has been hypothesized that an alternate and novel function exists for this system in the dissemination of genetic elements, notably antibiotic resistance genes, in Acinetobacter species. Currently the mechanism underlying this apparent genetic mobility is unknown. Multidrug resistant Acinetobacter baumannii is an increasingly problematic pathogen that can cause recurring infections. Sequencing of numerous plasmids from clinical isolates of A. baumannii revealed the presence of possible mobile modules: genes were found flanked by pairs of Xer recombination sites, called plasmid‐dif (pdif) sites. These modules have been identified in multiple otherwise unrelated plasmids and in different genetic contexts suggesting they are mobile elements. In most cases, the pairs of sites flanking a gene (or genes) are in inverted repeat, but there can be multiple modules per plasmid providing pairs of recombination sites that can be used for inversion or fusion/deletion reactions; as many as 16 pdif sites have been seen in a single plasmid. Similar modules including genes for surviving environmental toxins have also been found in strains of Acinetobacter Iwoffi isolated from permafrost cores; this suggests that these mobile modules are an ancient adaptation and not a novel response to antibiotic pressure. These modules bear all the hallmarks of mobile genetic elements, yet, their movement has never been directly observed to date. This review gives an overview of the current state of this novel research field.
中文翻译:

不动杆菌中 pdif 模块的动员:抗生素抗性基因改组的新机制?
XerCD - dif位点特异性重组是一个特征很好的系统,在大多数细菌和古细菌中都有发现。它的作用是解析同源重组产生的染色体二聚体。Xer 介导的重组也被一些质粒用于多聚体分辨率以提高稳定性,并被一些噬菌体用于整合到染色体中。在过去的十年中,有人假设该系统在不动杆菌属物种的遗传元件(特别是抗生素抗性基因)的传播中存在替代的新功能。目前,这种明显的遗传流动性背后的机制尚不清楚。多重耐药鲍曼不动杆菌是一种越来越成问题的病原体,可导致反复感染。从临床分离大量质粒的测序鲍曼不动杆菌揭示的可能移动模块的存在:被发现的基因通过双Xer重组位点,称为质粒的侧翼DIF(对DIF)位点。这些模块已在多个其他不相关的质粒和不同的遗传背景中被鉴定出来,表明它们是可移动的元件。在大多数情况下,一个基因(或多个基因)侧翼的位点对是反向重复,但每个质粒可以有多个模块,提供可用于反向或融合/缺失反应的重组位点对;多达 16 p差异位点已在单个质粒中看到。在从永久冻土核心分离的Iwoffi 不动杆菌菌株中也发现了类似的模块,包括使环境毒素存活的基因;这表明这些移动模块是一种古老的适应性,而不是对抗生素压力的新反应。这些模块具有可移动遗传元件的所有特征,但迄今为止从未直接观察到它们的运动。本综述概述了这一新研究领域的现状。
更新日期:2020-06-28
中文翻译:

不动杆菌中 pdif 模块的动员:抗生素抗性基因改组的新机制?
XerCD - dif位点特异性重组是一个特征很好的系统,在大多数细菌和古细菌中都有发现。它的作用是解析同源重组产生的染色体二聚体。Xer 介导的重组也被一些质粒用于多聚体分辨率以提高稳定性,并被一些噬菌体用于整合到染色体中。在过去的十年中,有人假设该系统在不动杆菌属物种的遗传元件(特别是抗生素抗性基因)的传播中存在替代的新功能。目前,这种明显的遗传流动性背后的机制尚不清楚。多重耐药鲍曼不动杆菌是一种越来越成问题的病原体,可导致反复感染。从临床分离大量质粒的测序鲍曼不动杆菌揭示的可能移动模块的存在:被发现的基因通过双Xer重组位点,称为质粒的侧翼DIF(对DIF)位点。这些模块已在多个其他不相关的质粒和不同的遗传背景中被鉴定出来,表明它们是可移动的元件。在大多数情况下,一个基因(或多个基因)侧翼的位点对是反向重复,但每个质粒可以有多个模块,提供可用于反向或融合/缺失反应的重组位点对;多达 16 p差异位点已在单个质粒中看到。在从永久冻土核心分离的Iwoffi 不动杆菌菌株中也发现了类似的模块,包括使环境毒素存活的基因;这表明这些移动模块是一种古老的适应性,而不是对抗生素压力的新反应。这些模块具有可移动遗传元件的所有特征,但迄今为止从未直接观察到它们的运动。本综述概述了这一新研究领域的现状。











































 京公网安备 11010802027423号
京公网安备 11010802027423号