当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Repurposing drugs against the main protease of SARS-CoV-2: mechanism-based insights supported by available laboratory and clinical data.
Molecular Omics ( IF 3.0 ) Pub Date : 2020-06-26 , DOI: 10.1039/d0mo00057d Sohini Chakraborti 1 , Sneha Bheemireddy , Narayanaswamy Srinivasan
Molecular Omics ( IF 3.0 ) Pub Date : 2020-06-26 , DOI: 10.1039/d0mo00057d Sohini Chakraborti 1 , Sneha Bheemireddy , Narayanaswamy Srinivasan
Affiliation
|
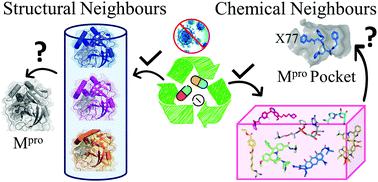
The ongoing global pandemic of COVID-19 has brought life to almost a standstill with the implementation of lockdowns and social distancing as some of the preventive measures in the absence of any approved specific therapeutic interventions. To combat this crisis, research communities worldwide are falling back on the existing repertoire of approved/investigational drugs to probe into their anti-coronavirus properties. In this report, we describe our unique efforts in identifying potential drugs that could be repurposed against the main protease of SARS-CoV-2 (SARS-CoV-2 Mpro). To achieve this goal, we have primarily exploited the principles of ‘neighbourhood behaviour’ in the protein 3D (workflow-I) and chemical 2D structural space (workflow-II) coupled with docking simulations and insights into the possible modes of action of the selected candidates from the available literature. This integrative approach culminated in prioritizing 29 potential repurpose-able agents (20 approved drugs and 9 investigational molecules) against SARS-CoV-2 Mpro. Apart from the approved/investigational anti-viral drugs, other notable hits include anti-bacterial, anti-inflammatory, anti-cancer and anti-coagulant drugs. Our analysis suggests that some of these drugs have the potential to simultaneously modulate the functions of viral proteins and the host response system. Interestingly, many of these identified candidates (12 molecules from workflow-I and several molecules, belonging to the chemical classes of alkaloids, tetracyclines, peptidomimetics, from workflow-II) are suggested to possess anti-viral properties, which is supported by laboratory and clinical data. Furthermore, this work opens a new avenue of research to probe into the molecular mechanism of action of many drugs, which are known to demonstrate anti-viral activity but are so far not known to target viral proteases.
中文翻译:

针对SARS-CoV-2的主要蛋白酶重新使用药物:现有实验室和临床数据支持的基于机制的见解。
在没有任何经过批准的特定治疗干预措施的情况下,作为一些预防措施,由于实行封锁和社会疏远,COVID-19的全球流行已使生命几乎停滞。为了应对这场危机,全世界的研究团体都在依靠现有的已批准/研究用药物目录来探究其抗冠状病毒特性。在本报告中,我们描述了我们为识别可能针对SARS-CoV-2(SARS-CoV-2 M pro)。为了实现这一目标,我们主要利用蛋白质3D(workflow-I)和化学2D结构空间(workflow-II)中的“邻居行为”原理,以及对接模拟和对选定对象可能的作用方式的洞察力现有文献中的候选人。这种综合方法最终确定了针对SARS-CoV-2 M pro的29种潜在可重用药物(20种已获批准的药物和9种研究性分子)的优先级。除了已批准/研究出的抗病毒药物外,其他值得注意的产品还包括抗菌,消炎,抗癌和抗凝药物。我们的分析表明,其中一些药物具有同时调节病毒蛋白和宿主反应系统功能的潜力。有趣的是,许多已鉴定的候选物(来自工作流程-I的12个分子和属于工作流程-II的生物碱,四环素,肽模拟物的化学类别的几个分子)建议具有抗病毒特性,这在实验室和实验室的支持下得以实现。临床资料。此外,这项工作为探索许多药物的分子作用机理开辟了新的研究途径,已知许多药物具有抗病毒活性,但迄今还不知道靶向病毒蛋白酶。
更新日期:2020-06-26
中文翻译:

针对SARS-CoV-2的主要蛋白酶重新使用药物:现有实验室和临床数据支持的基于机制的见解。
在没有任何经过批准的特定治疗干预措施的情况下,作为一些预防措施,由于实行封锁和社会疏远,COVID-19的全球流行已使生命几乎停滞。为了应对这场危机,全世界的研究团体都在依靠现有的已批准/研究用药物目录来探究其抗冠状病毒特性。在本报告中,我们描述了我们为识别可能针对SARS-CoV-2(SARS-CoV-2 M pro)。为了实现这一目标,我们主要利用蛋白质3D(workflow-I)和化学2D结构空间(workflow-II)中的“邻居行为”原理,以及对接模拟和对选定对象可能的作用方式的洞察力现有文献中的候选人。这种综合方法最终确定了针对SARS-CoV-2 M pro的29种潜在可重用药物(20种已获批准的药物和9种研究性分子)的优先级。除了已批准/研究出的抗病毒药物外,其他值得注意的产品还包括抗菌,消炎,抗癌和抗凝药物。我们的分析表明,其中一些药物具有同时调节病毒蛋白和宿主反应系统功能的潜力。有趣的是,许多已鉴定的候选物(来自工作流程-I的12个分子和属于工作流程-II的生物碱,四环素,肽模拟物的化学类别的几个分子)建议具有抗病毒特性,这在实验室和实验室的支持下得以实现。临床资料。此外,这项工作为探索许多药物的分子作用机理开辟了新的研究途径,已知许多药物具有抗病毒活性,但迄今还不知道靶向病毒蛋白酶。











































 京公网安备 11010802027423号
京公网安备 11010802027423号