当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Use of Extended-Hückel Descriptors for Rapid and Accurate Predictions of Conjugated Torsional Energy Barriers.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-06-26 , DOI: 10.1021/acs.jcim.0c00440 Wanlei Wei 1 , Candide Champion 2 , Stephen J Barigye 1 , Zhaomin Liu 1 , Paul Labute 2 , Nicolas Moitessier 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-06-26 , DOI: 10.1021/acs.jcim.0c00440 Wanlei Wei 1 , Candide Champion 2 , Stephen J Barigye 1 , Zhaomin Liu 1 , Paul Labute 2 , Nicolas Moitessier 1
Affiliation
|
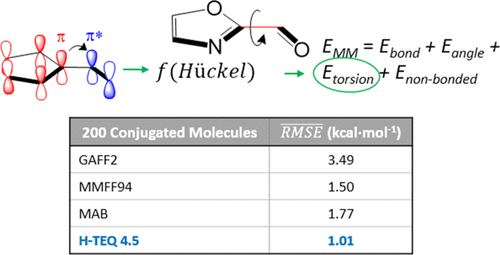
Over the past few decades, virtual high-throughput screening (vHTS) and molecular dynamics simulations have become effective and widely used tools in the initial stages of drug discovery efforts. These methods allow a great number of druglike molecules to be screened quickly and inexpensively. Unfortunately, however, the accuracies of both these methods rely on the quality of the underlying molecular mechanics force fields (FFs), which are often poor. This major weakness originates from the reliance of FFs on a finite list of specific parameters, called atom types, which have low transferability between molecules. In particular, the torsional energy barriers of druglike molecules are notoriously difficult to predict. Continuing our endeavor to understand factors affecting the torsional energy barriers of small molecules and quantify them, we showed that descriptors calculated using the extended-Hückel method could be used to rapidly assign accurate torsion parameters for conjugated molecules. This method, called H-TEQ 4.5, was developed using a set of 684 conjugated molecules. It was subsequently validated on a test set of 200 diverse molecules and produced an average root-mean-square error (rmse) of 1.01 kcal·mol–1, with respect to the reference quantum mechanic torsional profiles. For comparison, GAFF2, MMFF94, and MAB produced average rmse’s of 3.49, 1.50, and 1.77 kcal·mol–1, respectively. H-TEQ 4.5 is also computationally inexpensive, running just under 0.25 ms for a biphenyl molecule on a home computer, allowing it to be used for vHTS of large libraries of compounds. Overall, H-TEQ 4.5 solved the problems associated with the transferability of torsion parameters for conjugated molecules. This method was incorporated into the Molecular Operating Environment and will be available for a wide variety of applications.
中文翻译:

使用扩展的Hückel描述符快速准确地预测共轭扭转能垒。
在过去的几十年中,虚拟高通量筛选(vHTS)和分子动力学模拟已成为药物发现工作初期的有效且广泛使用的工具。这些方法允许快速且廉价地筛选大量药物样分子。但是,不幸的是,这两种方法的准确性都依赖于通常较差的潜在分子力学力场(FFs)的质量。此主要缺点来自FF对特定参数(称为原子类型)的有限列表的依赖,这些参数在分子之间具有较低的可转移性。特别地,众所周知,难以预测类药物分子的扭转能垒。继续努力了解影响小分子扭转能垒的因素并将其量化,我们表明,使用扩展的Hückel方法计算的描述符可用于快速分配共轭分子的准确扭转参数。此方法称为H-TEQ 4.5,是使用一组684个共轭分子开发的。随后在200个不同分子的测试集上对其进行了验证,得出的平均均方根误差(rmse)为1.01 kcal·mol–1,关于参考量子力学扭转轮廓。为了进行比较,GAFF2,MMFF94和MAB的平均均方根值分别为3.49、1.50和1.77 kcal·mol –1。H-TEQ 4.5在计算上也很便宜,对于家用计算机上的联苯分子而言,运行时间不到0.25毫秒,因此可用于大型化合物库的vHTS。总体而言,H-TEQ 4.5解决了与共轭分子扭转参数的可传递性相关的问题。该方法已整合到分子操作环境中,可用于多种应用。
更新日期:2020-07-27
中文翻译:

使用扩展的Hückel描述符快速准确地预测共轭扭转能垒。
在过去的几十年中,虚拟高通量筛选(vHTS)和分子动力学模拟已成为药物发现工作初期的有效且广泛使用的工具。这些方法允许快速且廉价地筛选大量药物样分子。但是,不幸的是,这两种方法的准确性都依赖于通常较差的潜在分子力学力场(FFs)的质量。此主要缺点来自FF对特定参数(称为原子类型)的有限列表的依赖,这些参数在分子之间具有较低的可转移性。特别地,众所周知,难以预测类药物分子的扭转能垒。继续努力了解影响小分子扭转能垒的因素并将其量化,我们表明,使用扩展的Hückel方法计算的描述符可用于快速分配共轭分子的准确扭转参数。此方法称为H-TEQ 4.5,是使用一组684个共轭分子开发的。随后在200个不同分子的测试集上对其进行了验证,得出的平均均方根误差(rmse)为1.01 kcal·mol–1,关于参考量子力学扭转轮廓。为了进行比较,GAFF2,MMFF94和MAB的平均均方根值分别为3.49、1.50和1.77 kcal·mol –1。H-TEQ 4.5在计算上也很便宜,对于家用计算机上的联苯分子而言,运行时间不到0.25毫秒,因此可用于大型化合物库的vHTS。总体而言,H-TEQ 4.5解决了与共轭分子扭转参数的可传递性相关的问题。该方法已整合到分子操作环境中,可用于多种应用。










































 京公网安备 11010802027423号
京公网安备 11010802027423号