Molecular Cell ( IF 14.5 ) Pub Date : 2020-06-26 , DOI: 10.1016/j.molcel.2020.06.007 Leina Lu 1 , Xiaoxiao Liu 1 , Wei-Kai Huang 2 , Paola Giusti-Rodríguez 3 , Jian Cui 1 , Shanshan Zhang 1 , Wanying Xu 1 , Zhexing Wen 4 , Shufeng Ma 5 , Jonathan D Rosen 6 , Zheng Xu 7 , Cynthia F Bartels 1 , Riki Kawaguchi 8 , Ming Hu 9 , Peter C Scacheri 1 , Zhili Rong 10 , Yun Li 7 , Patrick F Sullivan 11 , Hongjun Song 12 , Guo-Li Ming 13 , Yan Li 14 , Fulai Jin 15
|
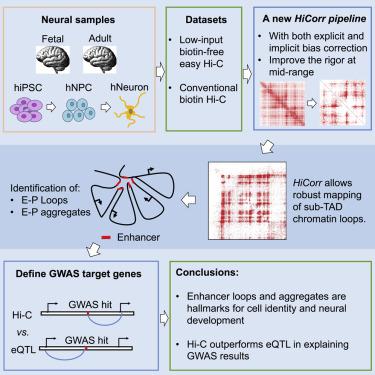
Genome-wide mapping of chromatin interactions at high resolution remains experimentally and computationally challenging. Here we used a low-input “easy Hi-C” protocol to map the 3D genome architecture in human neurogenesis and brain tissues and also demonstrated that a rigorous Hi-C bias-correction pipeline (HiCorr) can significantly improve the sensitivity and robustness of Hi-C loop identification at sub-TAD level, especially the enhancer-promoter (E-P) interactions. We used HiCorr to compare the high-resolution maps of chromatin interactions from 10 tissue or cell types with a focus on neurogenesis and brain tissues. We found that dynamic chromatin loops are better hallmarks for cellular differentiation than compartment switching. HiCorr allowed direct observation of cell-type- and differentiation-specific E-P aggregates spanning large neighborhoods, suggesting a mechanism that stabilizes enhancer contacts during development. Interestingly, we concluded that Hi-C loop outperforms eQTL in explaining neurological GWAS results, revealing a unique value of high-resolution 3D genome maps in elucidating the disease etiology.
中文翻译:

增强子-启动子相互作用的稳健 Hi-C 图谱揭示了非编码基因组在神经发育和疾病中的功能。
高分辨率染色质相互作用的全基因组图谱在实验和计算上仍然具有挑战性。在这里,我们使用低输入“easy Hi-C”协议来绘制人类神经发生和脑组织中的 3D 基因组架构,并证明严格的 Hi-C 偏差校正管道 ( HiCorr ) 可以显着提高亚 TAD 水平的 Hi-C 环识别,尤其是增强子-启动子 (EP) 相互作用。我们使用HiCorr比较了 10 种组织或细胞类型的染色质相互作用的高分辨率图,重点关注神经发生和脑组织。我们发现动态染色质环比区室转换更能成为细胞分化的标志。 HiCorr允许直接观察跨越大邻域的细胞类型和分化特异性 EP 聚集体,这表明了一种在发育过程中稳定增强子接触的机制。有趣的是,我们得出结论,Hi-C 环在解释神经 GWAS 结果方面优于 eQTL,揭示了高分辨率 3D 基因组图谱在阐明疾病病因学方面的独特价值。











































 京公网安备 11010802027423号
京公网安备 11010802027423号