Cell Systems ( IF 9.0 ) Pub Date : 2020-06-26 , DOI: 10.1016/j.cels.2020.05.010 Nicholas J Bernstein 1 , Nicole L Fong 1 , Irene Lam 1 , Margaret A Roy 1 , David G Hendrickson 1 , David R Kelley 1
|
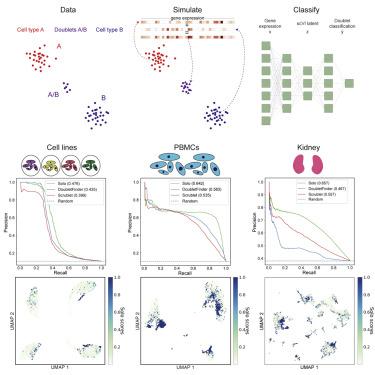
Single-cell RNA sequencing (scRNA-seq) measurements of gene expression enable an unprecedented high-resolution view into cellular state. However, current methods often result in two or more cells that share the same cell-identifying barcode; these “doublets” violate the fundamental premise of single-cell technology and can lead to incorrect inferences. Here, we describe Solo, a semi-supervised deep learning approach that identifies doublets with greater accuracy than existing methods. Solo embeds cells unsupervised using a variational autoencoder and then appends a feed-forward neural network layer to the encoder to form a supervised classifier. We train this classifier to distinguish simulated doublets from the observed data. Solo can be applied in combination with experimental doublet detection methods to further purify scRNA-seq data to true single cells. It is freely available from https://github.com/calico/solo. A record of this paper’s transparent peer review process is included in the Supplemental Information.
中文翻译:

Solo:通过半监督深度学习在单细胞 RNA-Seq 中识别双峰。
基因表达的单细胞 RNA 测序 (scRNA-seq) 测量能够以前所未有的高分辨率查看细胞状态。然而,当前的方法通常会导致两个或多个细胞共享相同的细胞识别条形码;这些“双峰”违反了单细胞技术的基本前提,并可能导致错误的推论。在这里,我们描述了 Solo,这是一种半监督深度学习方法,它比现有方法更准确地识别双峰。Solo 使用变分自编码器在无监督的情况下嵌入细胞,然后将前馈神经网络层附加到编码器以形成有监督的分类器。我们训练这个分类器来区分模拟双峰和观察到的数据。Solo 可以与实验双联检测方法结合使用,以进一步将 scRNA-seq 数据纯化为真正的单细胞。它可以从 https://github.com/calico/solo 免费获得。本文的透明同行评审过程的记录包含在补充信息中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号